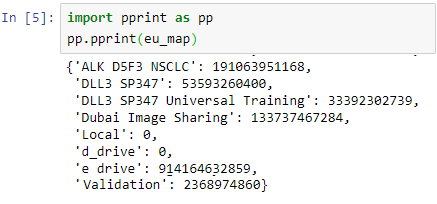
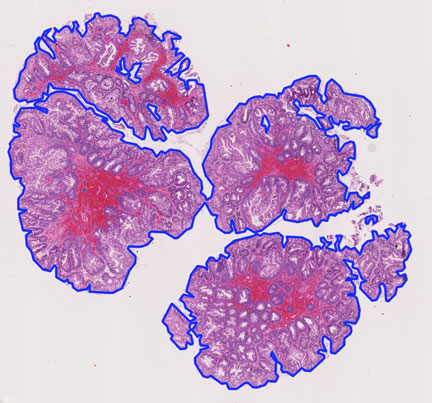
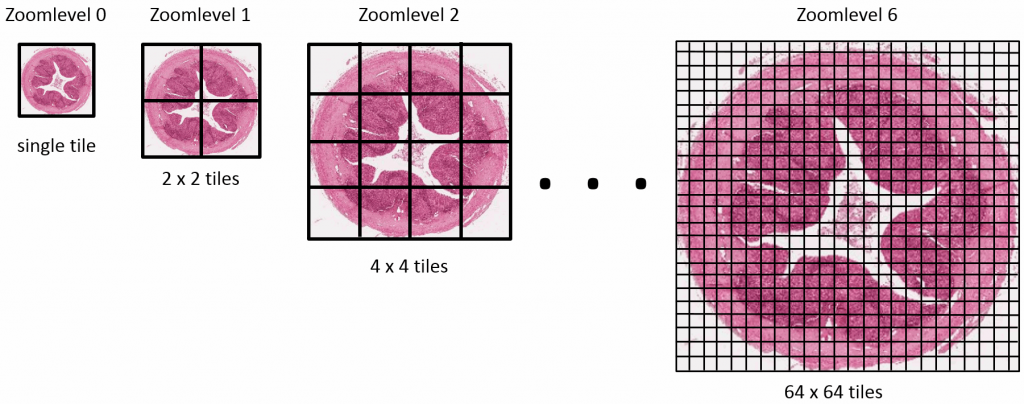
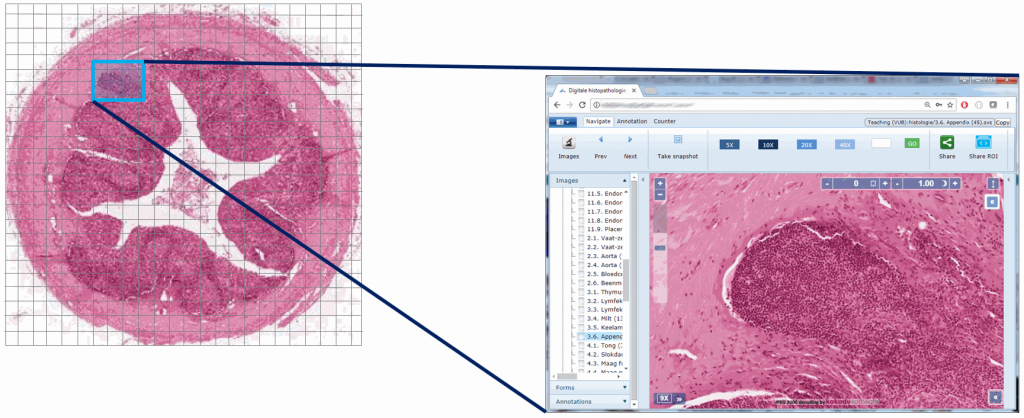
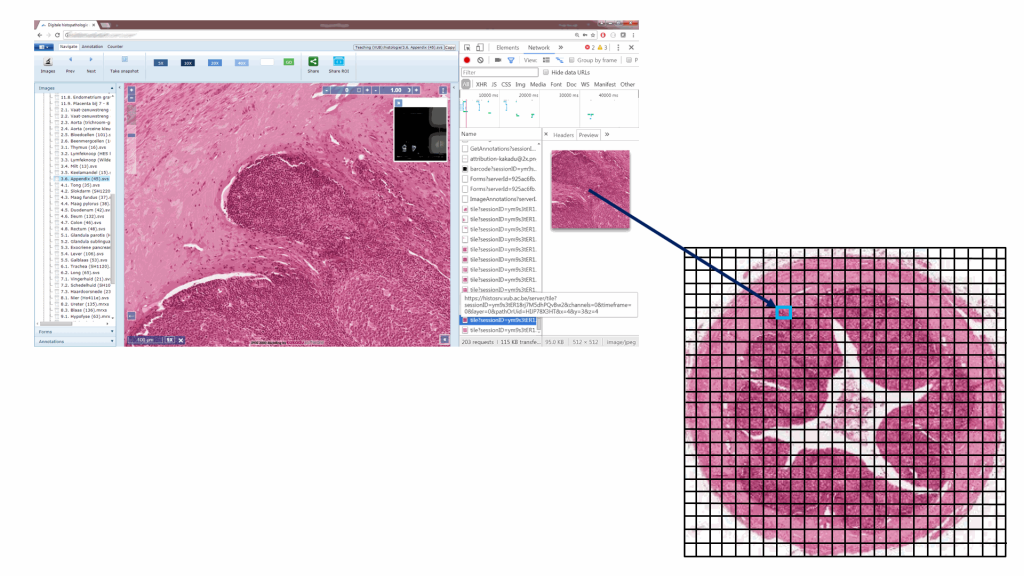
Educational needs and virtual microscopy
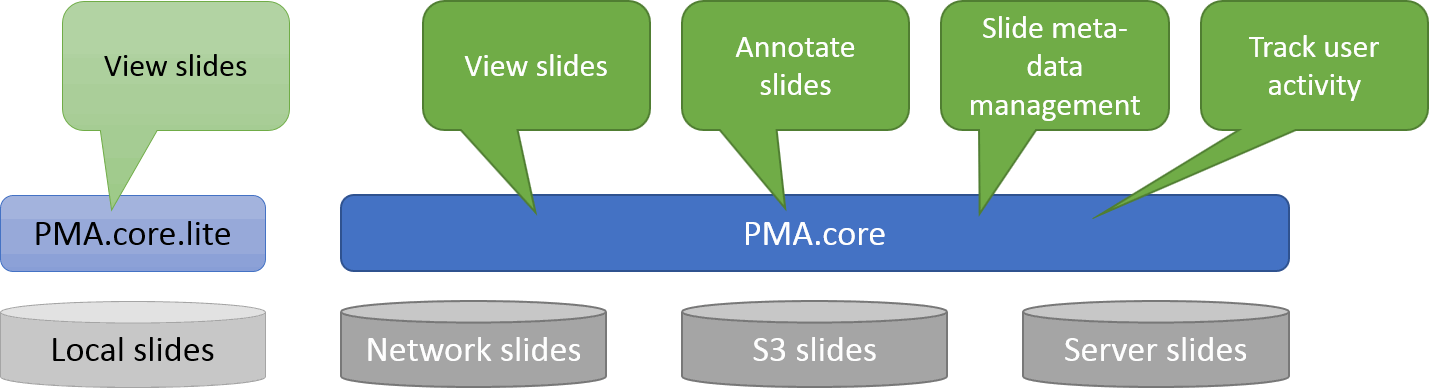
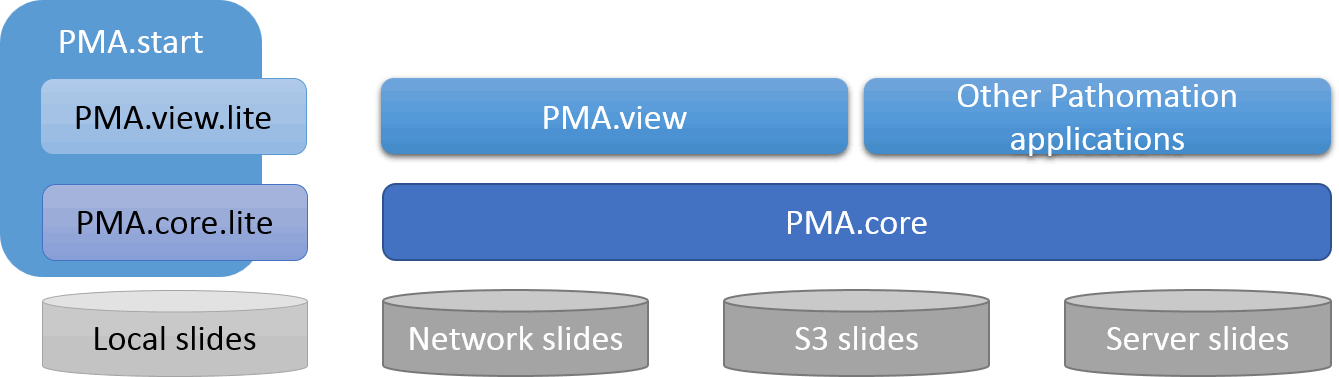
Pathomation’s software platform includes software for a number of digital pathology application.
At the most basic end of the spectrum we offer PMA.start for local viewing and research purposes. Even with PMA.start, you get full access to our front-end Javascript-based visualization framework, and back-end automation API (more on those in a separate blogpost).
At the opposite end of the spectrum, we offer a sophisticated training software package called PMA.control. Consider this:
- Current slide-based approaches to microscopy teaching face the logistical challenge of transporting people, slides and training material.
- The size, location and number of instructional sessions is limited (in time, place, and size)
- Concurrent training on the same material is not possible. One microscope can offer one unique slide. Musical chair… erh… microscopes, anyone?
We identified a need has evolved to train students and professionals alike to accurately evaluate tissue material with a broad range of (assay-specific) algorithms. Systematically organizing training materials and bringing training participants together in a virtual settings, is what it’s all about.
Projects
You start in PMA.control by setting up a project. A project describes what it is that you want to organize instructional material around. A project can represent a course at a university, or a drug for a pharma company. A project can delineate a geographical territory. It’s totally up to you.
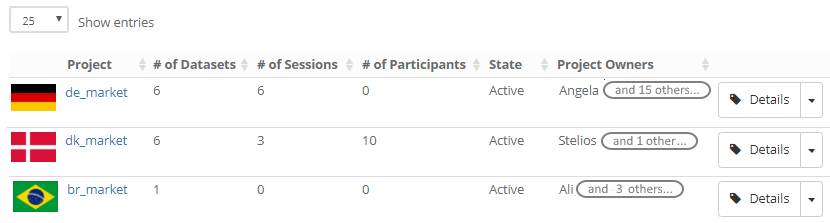
Projects have various properties. Apart from their name, you can identify them with an icon. This is convenient as your list of project becomes larger and more people become involved in your project. Speaking of involved people: you can identify one or more project managers. This is particularly useful for larger organizations, where one person is seldom available all the time, but it’s relatively easy to find a replacement in case of absence.
Training sessions
A project consists of training sessions. Again, what these mean semantically is completely up to you and your imagination:
- One client of ours uses PMA.control to organize weekly seminars in various places across the globe. Each seminar/country combination translates into its own training session in PMA.control, with specific start- and end-dates
- One medical school uses PMA.control to train residents. A training session can refer to the class coming in on a particular week, but it can also be linked to small research projects that students participate in.
- Another client integrates PMA.control into a web-portal, so all training sessions by definitions are open ended. The client has a drug portfolio, so rather than have them be restricted in time, training sessions refer to various indications for different drugs.
Safe to say that training sessions can be exploited for diverse applications.
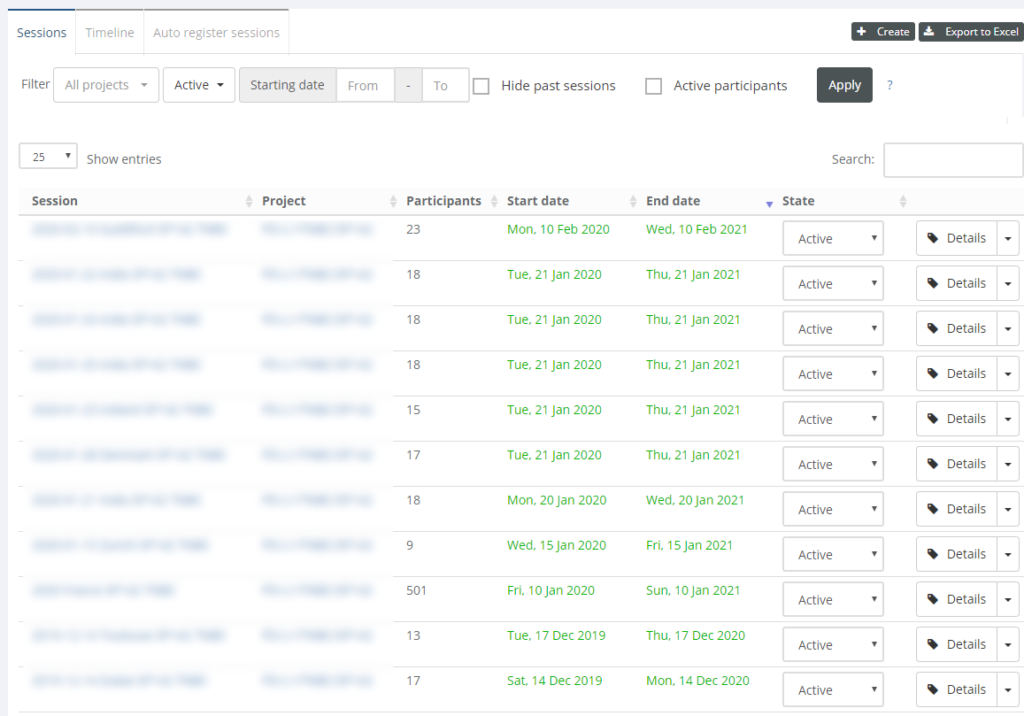
Case collections
All right, we have projects and training sessions… When do we put digital slide content in them? This is where case collections come in.
The idea is that you organize your training sessions in different parts. During a three-day seminar, you could have one day dedicated to guided lectures (that’s a case collection with its own slides). On day two, you allow people to evaluate themselves through some hands-on exercises (on a second case collection, which holds different slides than the first one). On day three, it’s crunch time, and attendees take an actual test to see how well they absorbed the material (on yet another third case collection with once again unique slides).
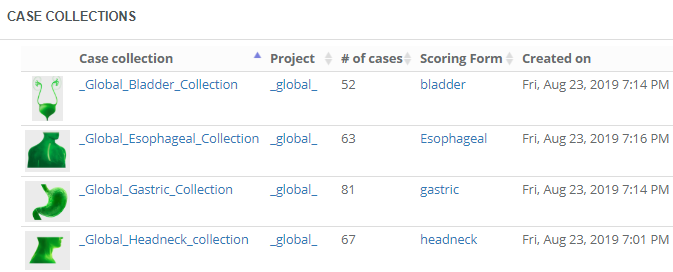
A case collection is coupled to a project, but is independent from any training sessions, so you can re-use them throughout the curriculum that you’re building. Think about it; otherwise if you organized the same training session repeatedly, you would continuously have to re-define the case collections, too!
A case collection consists of cases, which in turn consist of slides. You can choose to construct a case such that it pre-focuses on a particular region of interest (ROI) within a slide. You can also add various meta-data at case-level as well as slide-level. Not unimportantly: you can configure the initial rotation angle for each slide in the case. This is particularly relevant if your case consists of serial sections that may not be all in the exact same orientation.
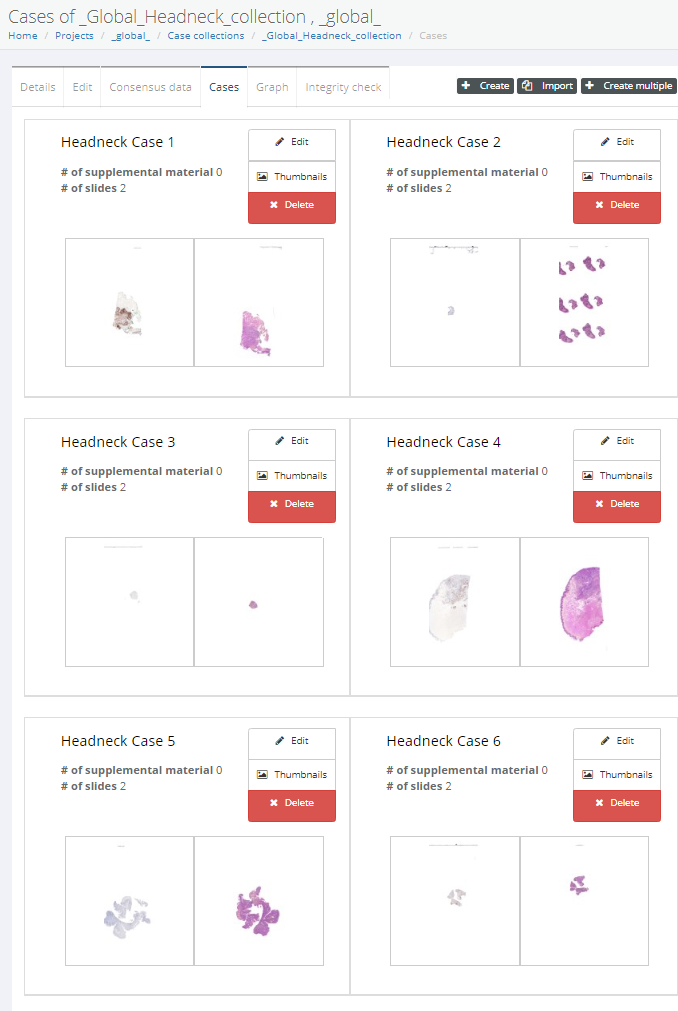
Interaction modes
Remember the three-day seminar we just mentioned? And you also remember that we called the software “PMA.control”, right?
Ok.
The name PMA.control refers to the fact that the owner of the software is in total control of what participants within a session at any given time.
Consider the following situations during our three-day seminar:
- On the first day, the instructor wants his pupils to stay nicely in the kiddie pool. They should give their undivided attention only to the material intended for the first day.
- However, this one person in the afternoon of the first day is taking the seminar for the second time. She asks if she can skip ahead already to the content from day two.
- On the second day, people are learning and experimenting with a different dataset. Do they understand the material well enough to pass the test on the last day? Clearly the material from day three must not be visible to anybody yet.
- On day three, it’s crunch time. The actual test material is now released. Depending on the intention of the instructor, earlier discussed material can now even be closed off.
For all these conditions, PMA.control offers interaction modes. An interaction mode controls if and how a case collection presents itself to the end-user.
Training sessions consist of multiple case collections and multiple users participate in a training session. At any given time, the instructor of a session can specify whether a particular case collection within the session is accessible to a specific user and how.
When signing into PMA.control, the instructor sees a grid with users and case collections. This grid can be used to control what user interacts with what case collection.
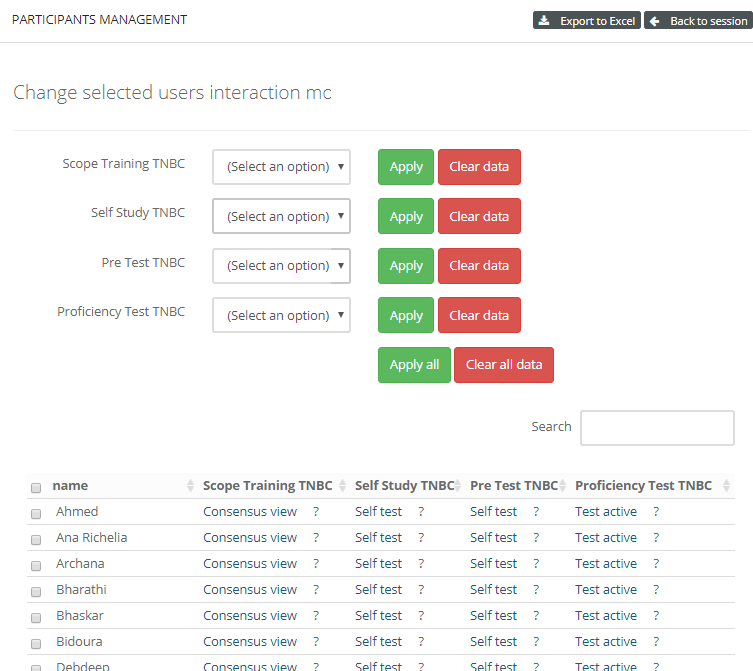
PMA.control ships with a number of default interaction modes, but these can be customized via a matrix interface where one stipulates what properties are associated with each.
Let’s see how interaction modes come into play during our three-day seminar:
- Before leaving for the seminar, the instructor applies the interaction mode “locked” to all case collections for all participants.
- On the morning of the first day, the instructor walks in the seminar room an hour early an sets the interaction mode of the first case collection to “browse” for everybody to see. That was easy! He goes to the hotel bar to grab a nice cup of coffee.
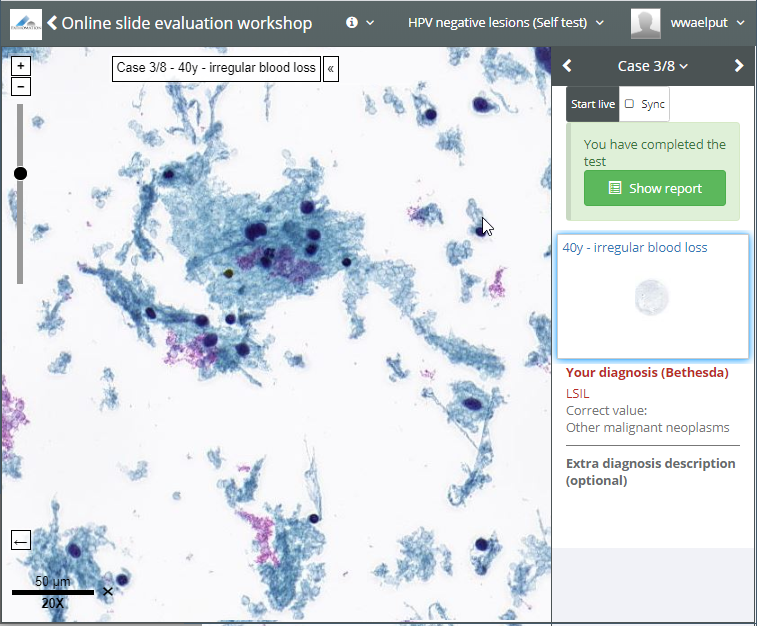
- In the afternoon of the first day, an attendee asks about being allowed to skip ahead with the material a bit. The instructor asks if any other people are in the same situation. For those, he sets the interaction mode for the second case collection to “self-test”. Users can interactively fill out a pre-determined scoring form that goes with each case, and they can see each other’s results to discuss their findings amongst themselves.
- On the second day, the second case collection is unlocked for everybody. Everybody now sees the second case collection in self-test mode. The first case collection remains in “browse” mode, so participants can use these as reference material. The third case collection remains off limits today.
- On the morning of the third day, the first thing the instructor does is reset the first and second case collection in the training session to “locked”. Rien ne va plus. The third and last case collection is switched to “test”, and students can take their final assessment. Users can interactively fill out a pre-determined scoring form that goes with each case, but they can’t see each other’s data anymore.
It’s possible for an instructor to be an instructor for one session, but only a “regular” participant in another. This is especially useful in medical schools where different specialists consult with and train each other on various subject matters on a continuous basis.
Conclusion
PMA.control allows you to assemble whole-slide images, scoring forms, consensus scores and scoring manuals into digital training modules. Full service, no-hassle, management of the software is offered through the PathoTrainer service, which is organized through Pathomation’s parent company CellCarta .

If you’re interested in higher education teaching of histology or pathology, make sure to also have a look at Pathomation’s education landing page. For pharma and CRO application, we have a separate landing page.