It’s coming, it’s coming, it’s coming, it’s coming…
And we’re really excited about it!
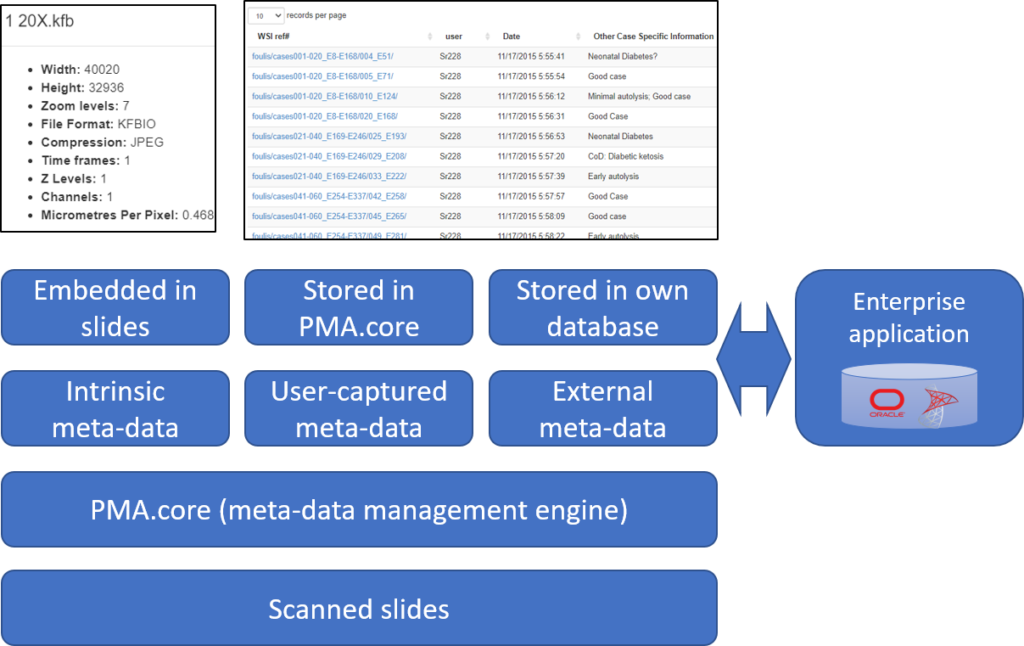
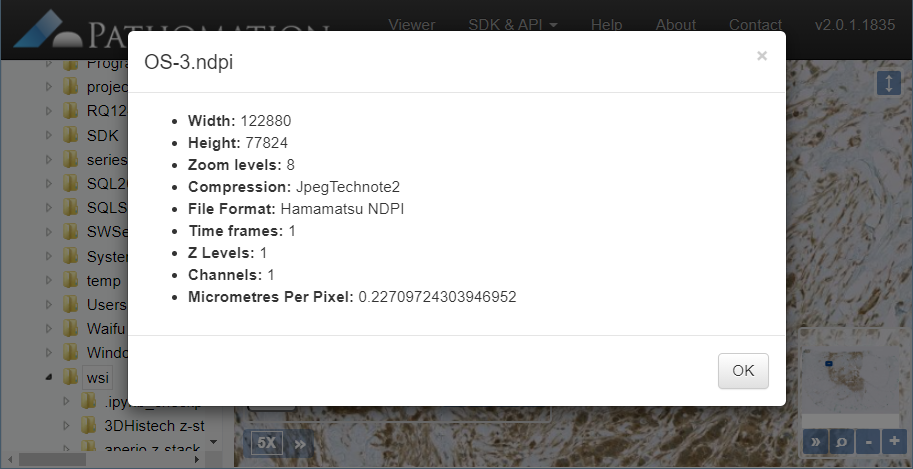
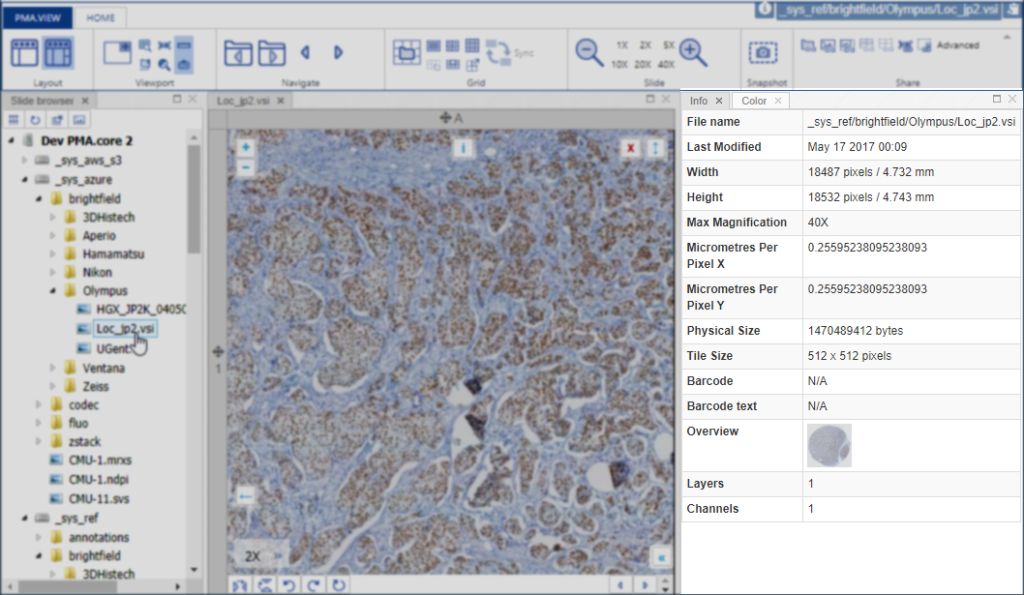
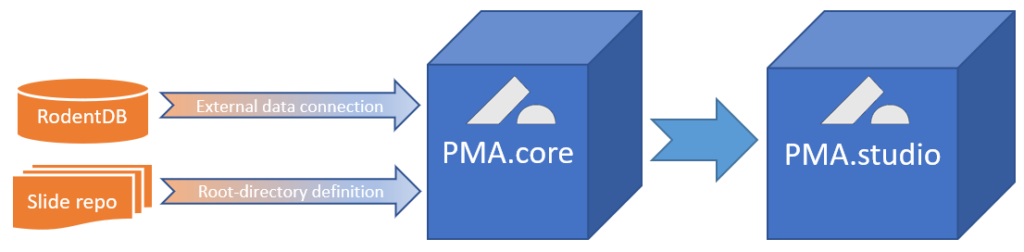
We’re in the process of wrapping up PMA.studio. This is going to be our flagship product for the next three years. It’s got everything a microscopy enthusiast needs, ranging from powerful viewport manipulation options, over annotations, to live conferencing. Remember our earlier article about how PMA.core handles (external) slide meta-data? All of that and more is in there, too.
In this blog post, we want to give developers as well as customers in our OEM and reseller program a sneak peek at PMA.studio and specifically focus on the opportunity for custom add-on development and white labeling.
PMA.studio is currently in testing phase for various use cases already, ranging from comparative validation studies to enterprise-wide deployment as a central histological information cockpit. If you are interested in joining the effort and helping us track down last minute bugs, do shoot us an email and we can see if you’re a good fit for our beta-program.
You can find more information about PMA.studio itself in the landing page that we’re building for the software at https://www.pathomation.com/pma.studio.
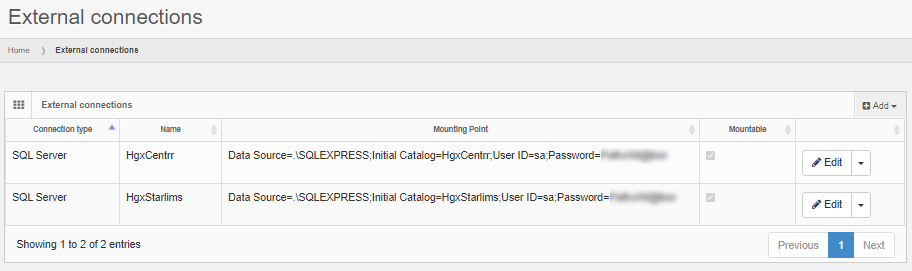
Administrative console
PMA.studio has a separate interface apart for administrative tasks.
In the administrative console, you can register any number of PMA.core instances to be used by PMA.studio to retrieve slides from. Once in use, for each registered PMA.core instance, you can see what users have sought access to it.
There’s a general dialog where you can set system-wide settings, like the address to contact in case of trouble, or the company / institute logo to use. This is where white labeling starts.
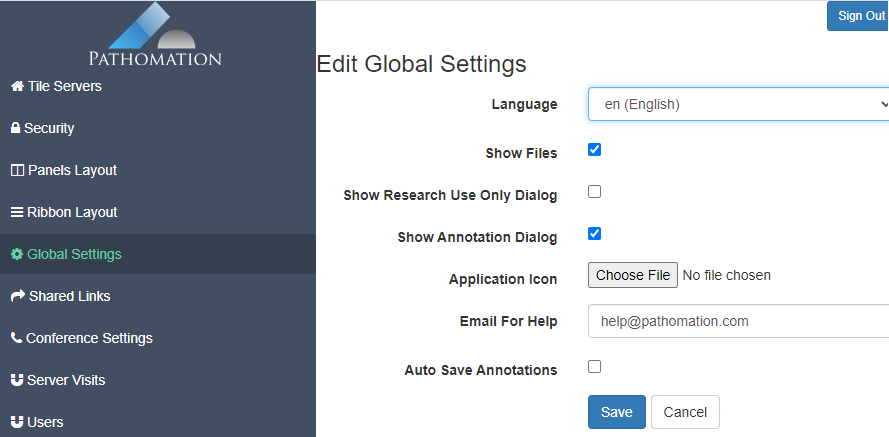
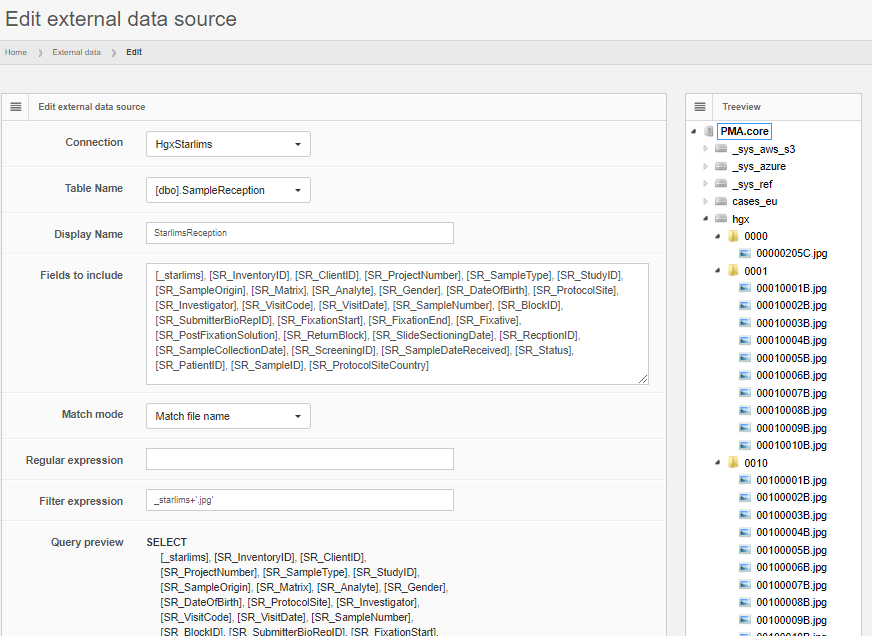
Like for PMA.view 1.x, the administrative console in PMA.studio can be used to customize the ribbon interface at top of the interface. The syntax is XML. We don’t have any formal definition of the format, and typically work with customers to get the result they want.
You can play around with the XML code yourself if you want. The default toolbar provides enough example code to allow simple re-arrangements of buttons and move features between different tabs.
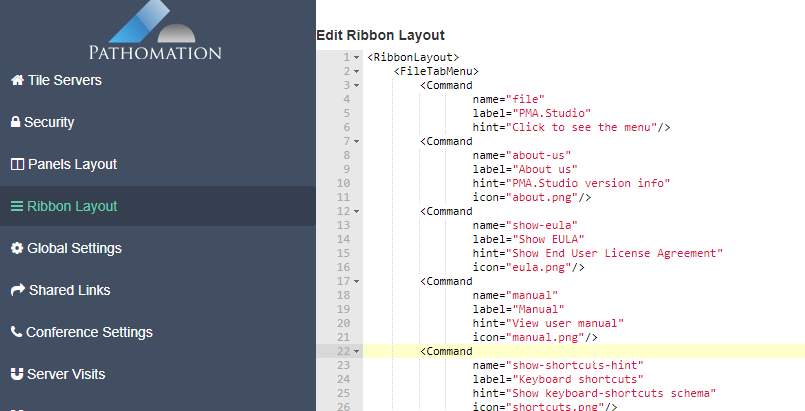
If you run into trouble, there’s always the option to restore the code back to a default toolbar.
Custom annotations
PMA.studio supports annotations. We have a separate tutorial on the subject as part of our beta-program.
There are situations where you don’t want users to go off and make their own annotations at will.
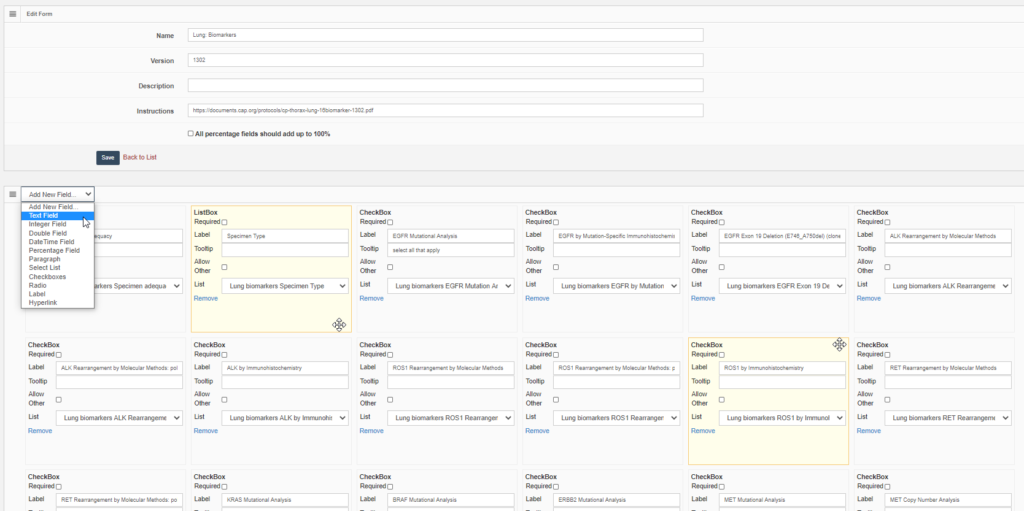
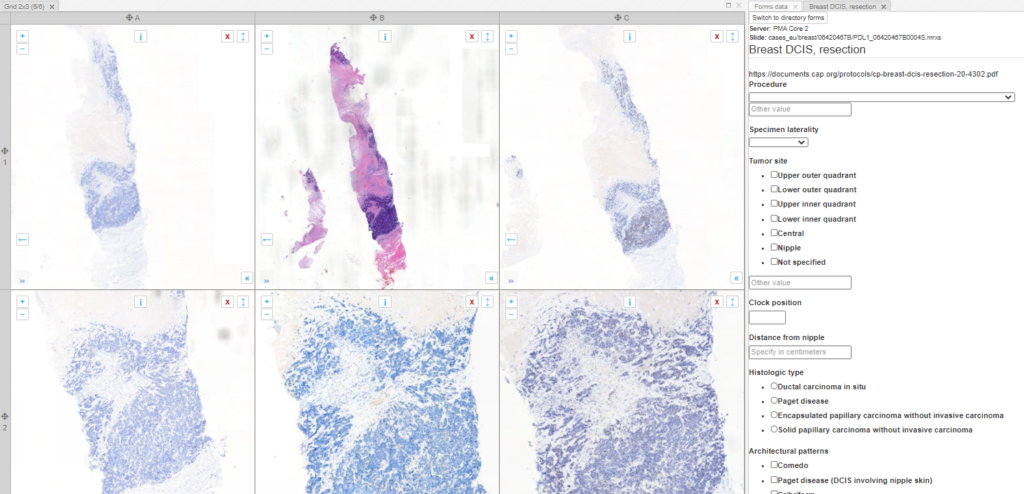
You may want your pathologists to annotate invasive tumor margins or necrotic areas for a study protocol. It’s useful then that people align, and everybody uses the same color scheme. So in addition to providing standard annotation tools, it’s possible to define buttons on your toolbars w/ pre-set attributes and parameters.
Here’s a great example of a project we participated in recently, for which participants had to indicate six different types of tissues / cells:

The underlaying XML for this extra ribbon tab is as follows:
<Tab label="NKI" hint="custom ribbon tab" name="nki_annotations" enabled="true" visible="true">
<Ribbon label="Total TLS" width="30%">
<Tool type="buttons" size="large">
<Command name="preset-peri_mat" icon="an_m_peri_tls.png" hint="Total # mature TLS (peritumoral)" label="Peritumoral, mature"
pma-classification="peri mature"
pma-shape="ClosedFreehand"
pma-color="#ffff33"
/>
<Command name="preset-intra_mat" icon="an_m_intra_tls.png" hint="Total # mature TLS (intratumoral)" label="Intratumoral, mature"
pma-classification="Intra mature"
pma-shape="ClosedFreehand"
pma-color="#cccc00"
/>
<Command name="preset-peri_immat" icon="an_im_peri_tls.png" hint="Total # immature TLS (peritumoral)" label="Peritumoral, immature"
pma-classification="Peri immature"
pma-shape="ClosedFreehand"
pma-color="#999900"
/>
<Command name="preset-intra_immat" icon="an_im_intra_tls.png" hint="Total # immature TLS (intratumoral)" label="Intratumoral, immature"
pma-classification="Intra immature"
pma-shape="ClosedFreehand"
pma-color="#ffff66"
/>
</Tool>
</Ribbon>
<Ribbon label="Pre-sets" width="30%">
<Tool type="buttons" size="large">
<Command name="preset-necrosis" icon="an_necrosis.png" hint="Tumor" label="% necrosis"
pma-classification="Necrosis"
pma-shape="ClosedFreehand"
pma-color="#aa0000"/>
<Command name="preset-fibrosis" icon="an_macro.png" hint="Tissue" label="% fibrosis"
pma-classification="Fibrosis"
pma-shape="ClosedFreehand"
pma-color="#00aa00"/>
<Command name="preset-via_tumor" icon="an_tumorcells.png" hint="Tissue" label="% viable tumor cells"
pma-classification="Viable tumor cells"
pma-shape="ClosedFreehand"
pma-color="#000000"/>
</Tool>
</Ribbon>
</Tab>
The pay-off of this kind of configuration is two-fold: the protocols are consistently executed as intended. In addition, the training needed for novel users to interact with the software is reduced because they only have a few options to choose from. Less can definitely be more in these scenarios.
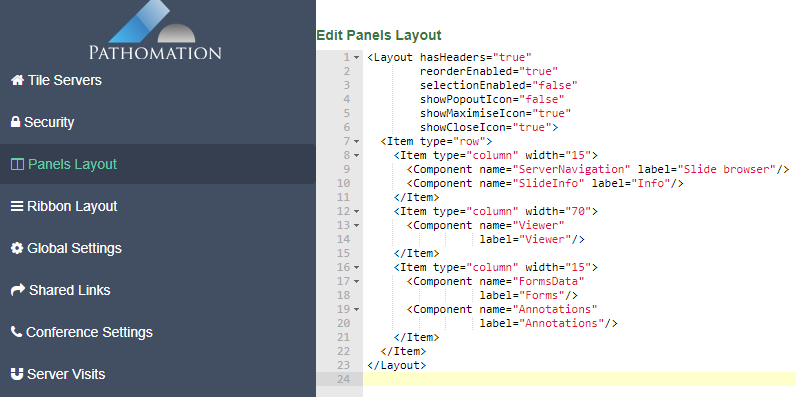
Configuring the panel layout
PMA.studio organizes its content across different panels. The layout of PMA.studio is a lot more powerful and flexible than in PMA.view 1.x.
All panels can be turned on or off via the Configuration tab on the ribbon. In addition, the admin console can be used to pre-determine the layout and panel organization when people first log in.
In its simplest configuration, you’re navigating slides and looking at them one by one:
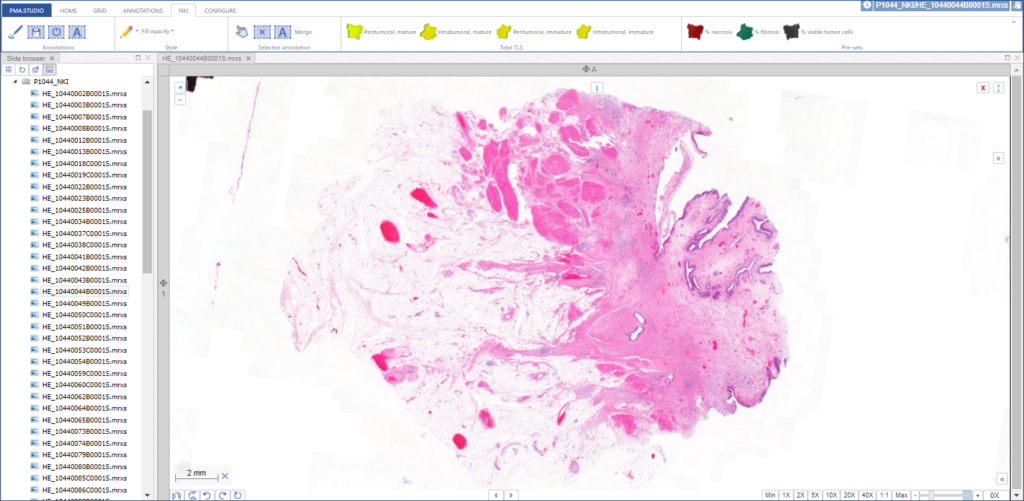
You can add various information panels as you see fit to get to something that looks more like this:
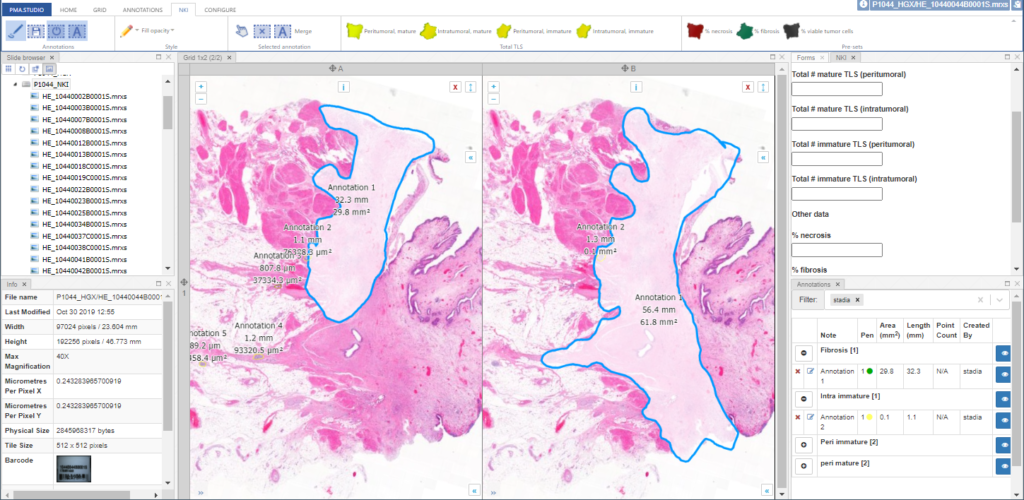
Like with the ribbon customization and pre-defined annotation controls, you can pre-set PMA.studio’s panel layout. Again, you’re looking at XML snippets.
The layout from the latter screenshot is then defined as follows:
Iframe panels
A special type of panel is the iframe-panel. With these panels, you can virtually load any website or page that you want within a pre-defined PMA.studio panel. You can define your own panel through the Layout configuration button on the ribbon:
A pop-up dialog appears and let’s you specify the title of the panel, and the URL you want it to load.
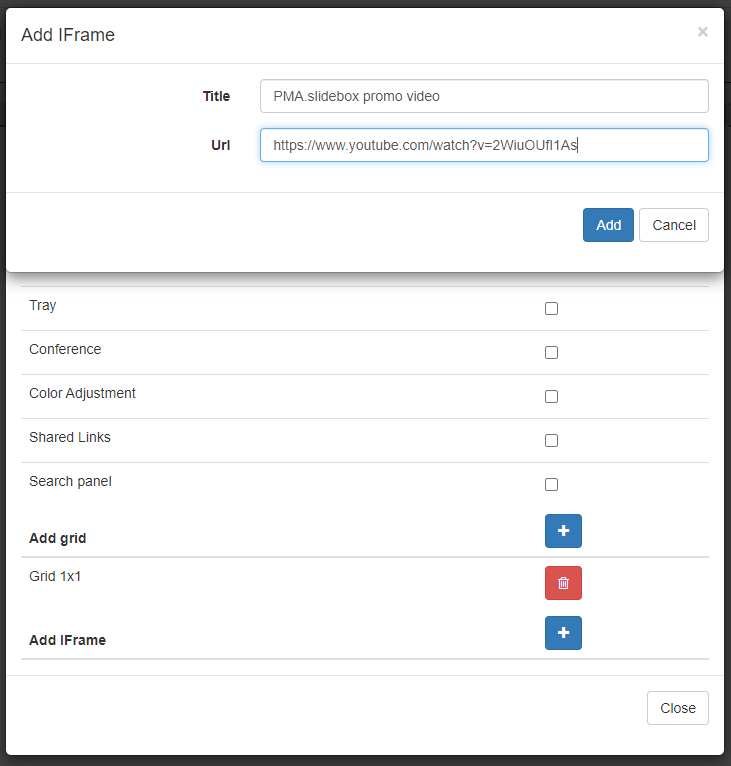
In our example we took a promotional video for our own PMA.slidebox product (more on that later), but you can put just about anything in there.
The result looks like this:
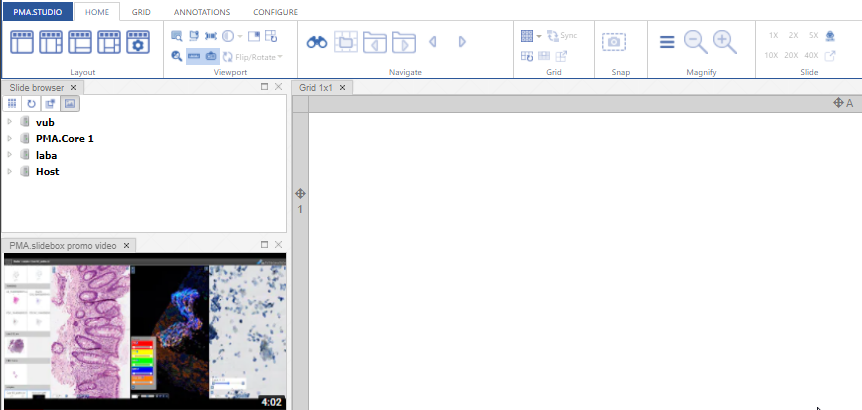
You can pre-define these custom iframe-panels any way you want through the admin console.
<Component name="IFrame" label="Pathomation video" url="https://www.youtube.com/embed/VsiGz8ykuEo"/>
Respective parameters that indicate the current state of the active viewport in PMA.studio are passed along automatically, like this:
http://www.server.com/app/page.php
?server=%PATH%TO%PMA%CORE%
&path=%FULL%PATH%OF%SLIDE%
&sessionId=%SESSION%ID%
Last but not least: a product pitch
We wrote earlier about PMA.slidebox.
This week, we’ve officially started promoting this product. We updated the landing page for PMA.slidebox, and the first demonstration videos are available through our YouTube channel.

Like with everything we do, we’re very excited about being able to package a ton of useful functionality in nevertheless compact product offering. Do have a look at our demo portal for PMA.slidebox and let us know if this something you, too, are interested in, too!