Evil money-grabbing thieves
Pathomation is a for-profit company. We pride ourselves in offering various commercial products. Over the years, we’ve developed a comprehensive software platform for digital pathology and virtual microscopy. This platform consists of various components, which can be purchased, rented, or even licensed.
We strive to contribute to the Open Source community where we can. Some of our developers get referenced occasionally in papers, and our SDKs are publicly available through GitHub. On occasion, we contribute to Open Source projects ourselves.
Sample slides
Occasionally, people also ask us about where to find sample slides.
A great inventory of publicly available whole slide images can be found at both OpenSlide and the OMERO project:
What else do you need?
Oh yes, a way to actually read these data!
Reading WSI data
There are two well established Open Source projects for WSI data and virtual microscopy:
- https://www.openslide.org
- https://www.openmicroscopy.org (OMERO / BioFormats)
As you’re getting started adding digital pathology features to your own software, there are other routes to explore, too:
- Work with slide scanner vendors
- Adapt or translate code from BioFormats
- Adapt or translate code from OpenSlide
All of these have downsides however:
- Scanner vendors: You can start negotiating with any particular scanner vendor to adapt their proprietary SDK. Congrats; after months of lobbying work, you’ll have the (rather unpleasant) job of tasking one of your engineers to incorporate a new arcane DLL into your code, for a single framework (so expect to repeat this process over and over again for the next couple of years as your platform gains broader adoption)
- Bioformats: is open source, so you could learn from their code and translate parts of it into your own. You can try it at least. Bioformats is a Java stack. If you can integrate that directly, you’re lucky. You could possibly transcribe the original Java-code to your own platform of course (C#, C++, Python, Rust, Haskell, Martian…). Same thing here: you’ll have the (still rather unpleasant, IMHO) task of tasking one of your engineers to actually do all of this, for a single file format (and again expect to repeat this process over and over again for the next couple of years as your platform gains broader adoption)
- OpenSlide: lots of support, lots of bindings, but a bit outdated (limited file format support), limited functionality (not a server, no concept of “tiles”, …), and not adapted for today’s regulatory environment (GAMP, CE-IVD(R), 21CFR.11, …).
Of course all of this might change in the future. But as you sit and wait, we are here to help now, and you may already consider what Pathomation has to offer as an alternative:
PMA.start
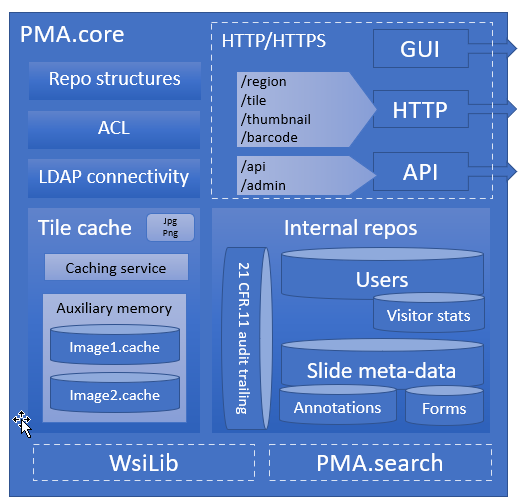
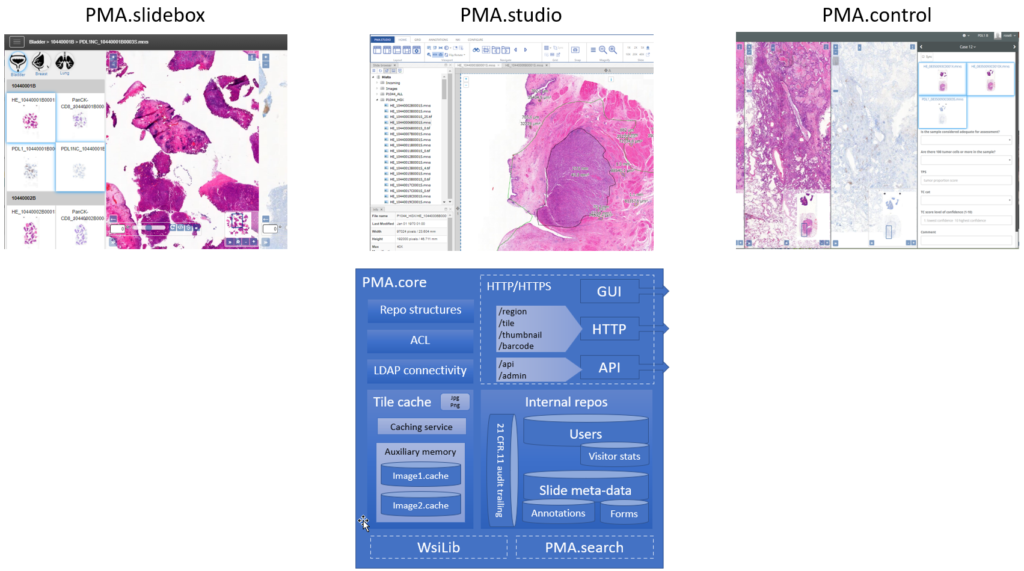
A central theme in our platform is the PMA.core tile server. A light-weight version of PMA.core is available free of change as PMA.start through https://free.pathomation.com.
PMA.start (and the entire Pathomation software stack by extension) supports any number of native file formats. So, you have to choices to go with here actually:
- Run our DICOMizer tool to convert the MRXS slides into DICOM data; see https://realdata.pathomation.com/wsi-digital-pathology-dicom-wg26/
- Script PMA.start to convert the slides into TIFFs; see our paragraph on libvips at https://realdata.pathomation.com/blur-detection-in-z-stacks/
The advantage of this approach? Once you’re in the DICOM or TIFF world, you’re pretty much independent from any proprietary software vendor. Use Paint.Net to make snapshots of particular Regions of Interest (ROIs), use Photoshop to run a sequence of filters to detect hormone receptors in breast biopsies.
The downside: this doesn’t scale particularly well with large datasets or high throughput.
It is our view then that reading native file formats directly will most likely remain the most efficient way to offer digital pathology services in the foreseeable future.
The hidden cost of free software
Our point with all the above is: free is not free. You can always download free software, but after 10 years I daresay it’s NEVER going to do what you actually need it to do. But you’ll need to invest R&D, write your own unit and integration tests, add additional regulatory and compliance layers, combine development stacks and frameworks (the proverbial round peg in square hole)… Assuming that you have these people in-house, or can find them: these DON’T work for free.
Last but not least, there’s an opportunity cost here as well: again, if you have super-dev-guy/girl; don’t you want him/her to work on the core mission of your company (like saving patients’ lives, or developing awesome eXplainable AI)?
Plan ahead if you can
We’re not against Open Source software. We don’t advocate a radical choice between the two. We think this can be an inclusive and/and story, where both models can live in harmony.
We do think however that in many scenarios, Pathomation is the right (and even the better) course of action from the start. Adapt PMA.start early on in your projects as an individual single user solution. Then, as you learn more and expand the scope of your application, talk to us on how to scale up and how to adapt PMA.core as a tile server in your enterprise architecture or organization.
We can resolve the file format handling problem once and for all _for_ you, so you have more time and energy to focus on your _actual_ application.
Even in the early stages; the limited cost that comes with our commercial solution and proper implementation support hugely outweighs the incurred costs from trying to tackle this middleware problem yourself.