On-slide annotations
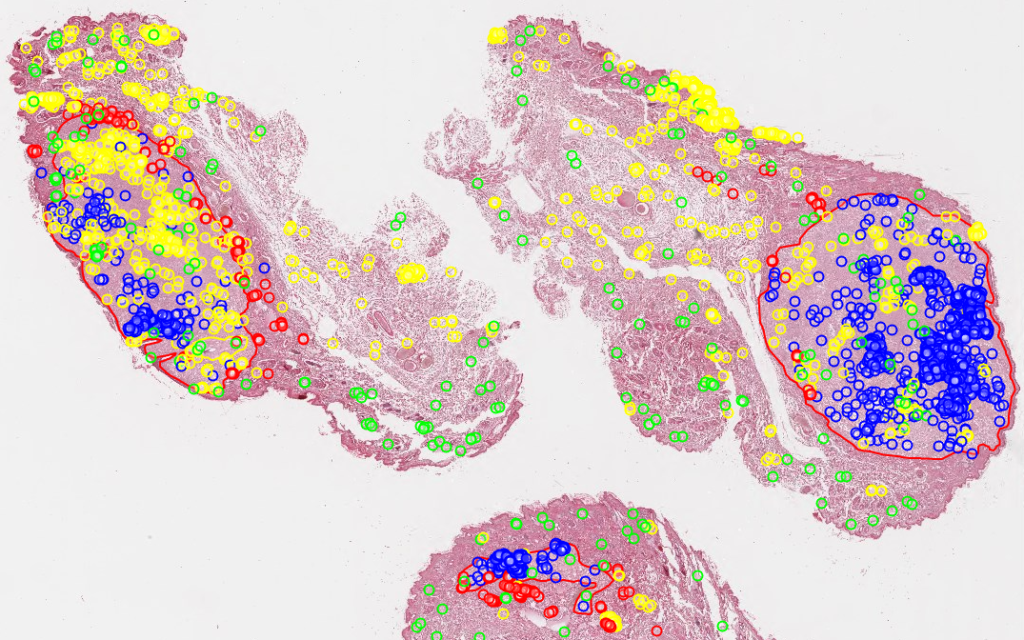
Do you recognize the above image? It’s a rendering of the mitotic figures and algorithmic classifications as determined by Bertram at al in their 2019 paper.
Digital pathology and virtual microscopy are concerned with slides (duh), but those are only part of the equation. External data as well as on-slide annotations are an integral part of the package, and any self-respecting software in this space supports various flavors of annotations. The various components in the Pathomation platform are no exception.
First some terminology: we generally refer to on-slide annotations that are geometric shapes and figures presented on top of virtual slide pixels in order to distinguish from (text-based) slide meta-data. The latter is presented as data attached to a slide, but not particularly or directly associated with a specific region on the slide. We discussed how Pathomation handles various flavors of slide metadata in an earlier blog post.
Creating on-slide annotations in PMA.studio
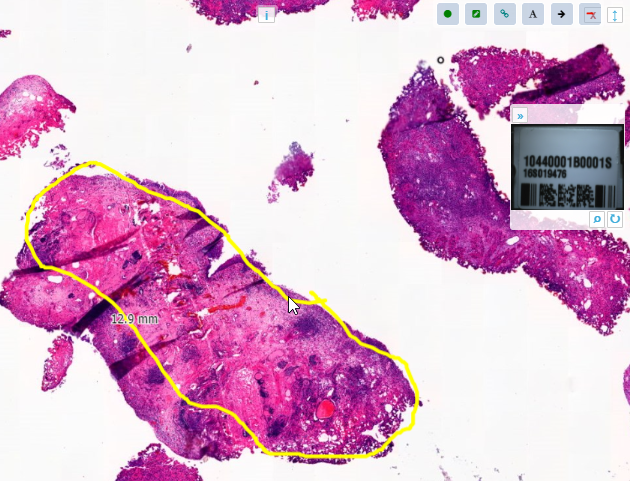
With the forthcoming release of PMA.studio 2.0, you can create on-slide annotations interactively.
PMA.studio offers a ribbon tab for this purpose. The first group of buttons lets you control what exactly is visible: the annotations themselves can be toggled, and you can choose whether to include the annotations labels or not.

The Style group in the ribbon is used to change the presentation of annotations. Annotations can have an edge color, fill color, which can be set independently from each other. Filled shapes can have a transparency attribute, too.
Each annotation can be associated with a class and a description attribute. An example would be a set of polygon shapes that each indicate necrosis and therefore have the “necrosis” class attribute, while individual shapes would be referred as a “necrosis-region-1”, “necrosis-region-2” etcetera.
Once you’ve made the annotations, you can follow-up on them through the annotations panel. Here, annotations are grouped per class. You can also use this panel to filter annotations per user.
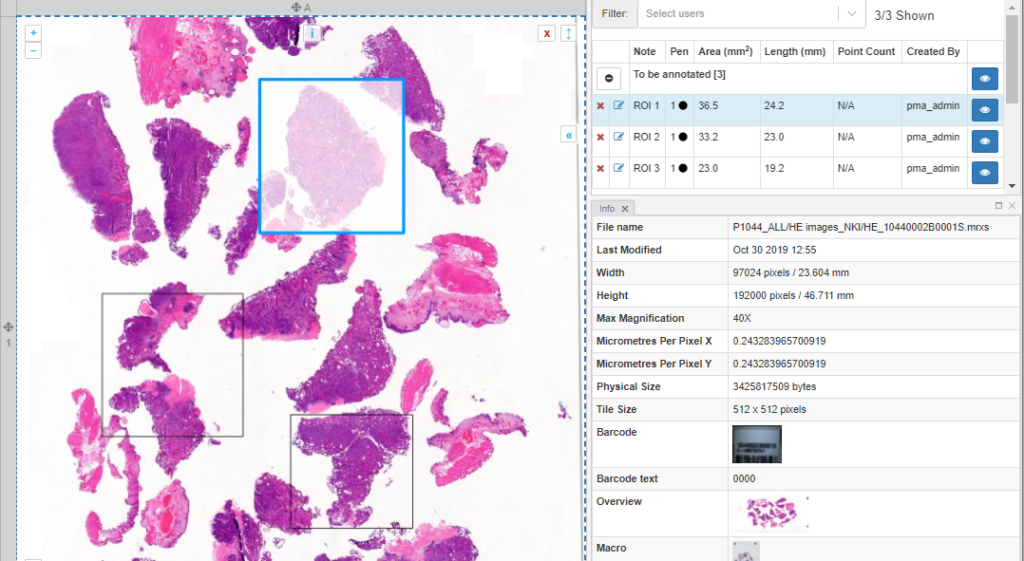
PMA.studio’s ribbon can be customized. This means that custom annotations are possible, too. You can use this feature to implement protocols.
In the example below, we’ve used XML to define pre-sets for pathologists to indicate various types of TLS regions:

Behind the scenes sits PMA.core
Let’s spend some time on how it all works behind the scenes.
When you make annotations in PMA.studio, you interact with a PMA.UI viewport. Once you decide to save you annotations, it’s the PMA.UI viewport that sends your annotations to PMA.core, where they are saved in the back-end database.
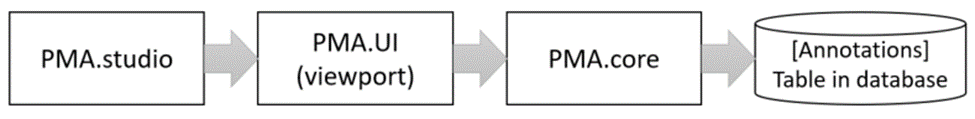
The format in which annotations are saved is Well-Known Text (WKT).
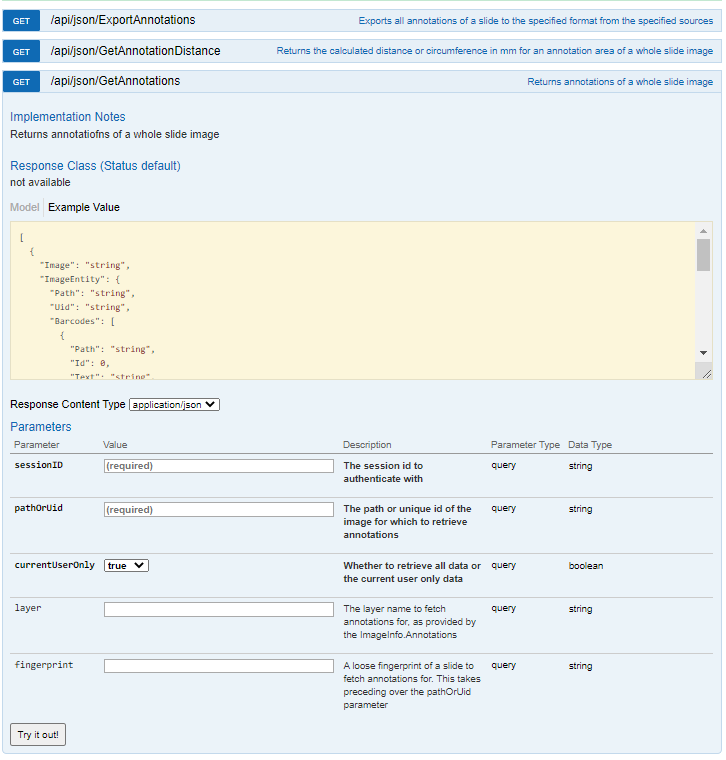
PMA.core has API calls to work with annotations. PMA.UI makes heavy use of these.
External annotations
Because we totally understand that PMA.studio may not be your first environment of choice to make your on-slide annotations with (though we think it’s a really good one!), PMA.core supports various other types of on-slide annotations, too.
We distinguish between “native” and “third-party” annotations.

Native annotations are shapes and forms included in the original vendor’s file format. Several vendors provide the option of making on-slide annotations in their own native viewers. Examples include 3DHistech and Aperio. If you have a 3Dhistech MRXS file that has overlaying annotations created by Case Center or the Pannoramic viewer, PMA.core will render them accordingly. The same goes for Aperio SVS files: just make sure you put the .xml file from ImageScope next to the corresponding .svs file.
Other vendors support annotations, too. If you find yourself with a vendor-specific annotation file format, do tell us, and we’ll add it to our next version of PMA.core.
Third-party annotations are another kind than “native” annotations. Like PMA.core’s own (WKT-encoded) annotations, these are created in software that is not coming from the vendor. Typically we’re talking about image analysis software. The three big names are out there are Definiens, Visiopharm, and Indica Labs HALO. Each of these environments is supported.
We’re not technically hindered to only support these abovementioned flavors of image analysis. So if you have a different environment from the one listed, let us know.
Transient annotations through PMA.live
Two products of Pathomation support real-time conferencing: PMA.studio and PMA.control. While in a conference, it is possible for participants to make live annotations that only exist for the duration of the conference.
Transient annotations in PMA.studio and PMA.control (through a third component referred to as PMA.live) can perhaps best be compared to the annotation toolbar that appears while giving a Powerpoint presentation: several tools are made available to temporarily highlight specific features, but these don’t become part of the original presentation.
Bringing it all together
As the “middleware for digital pathology and virtual microscopy” company, we take our job seriously. Therefore, apart from managing slides, we also allow our tile server PMA.core to be used to organize graphical annotations. The Pathomation platform offers many ways to organize
Pathomation can ingest both native and third-party annotations. The difference is that we consider native annotations to be annotations made with the original manufacturer’s (viewer) software, while third-party annotations are annotations created by independent vendors like Indica Labs or Visiopharm.
We also allow people to make annotations on top of slides using only components within the Pathomation platform. You don’t need external software to get started with annotating your slides; you can use our own PMA.studio, or even couple OpenCV output back to PMA.core through our API.
There’s more to say about annotations. In upcoming articles, we plan to show you how you can manage heterogenous annotation data from many sources, as well as how to use the back-end API directly to feed annotations back to PMA.core.
Want to see us work out a specific use case for your annotation workflows? Let us know.

