Image data types
Pathomation is concerned with any (imaging) data that is microscopy- or pathology-related. Much of the data is large: we talk about gigapixel data, or (more apt for microscopy) whole slide images (WSI).
Not all image data accessed via Pathomation need be large though:
- Microscopic images can represent individual fields of view, specific areas of interest captured by a mounted camera and for discussion or other ad hoc purposes
- Pathology starts from physical tissue. Therefore, it is often useful to photograph the obtained tissue. These are typically referred to as “macroscopic images”.
Oftentimes the above results can be stored in common (image) file formats: JPEG and TIFF are most often encountered. You can distribute these images via any medium. But if you have them side by side with your high-resolution images representing prepared slides, it’s reassuring to know that with Pathomation software you can organize these different slides under one umbrella.
Back to the really big data. Elsewhere on this blog, we have an article about the technical challenges when wanting to store an image that contains 100,000 x 200,000 pixels.
We send said article to (potential) customers regularly. Some are helped by it, some not. Because, understandably, many times you just want to know about your slides. When you book a flight, you don’t want to get an explanation about Newton’s or Bernouilli’s laws either… Just get me the tickets please.
So why did we write the article in the first place? Because at Pathomation, we pride ourselves at being format-agnostic. We don’t pick hardware vendors. Each scanner has their pros and cons. Each comes out to server a specific market segment and works better with some tissues than others. It’s not our place to subjectively decide who’s better or worse (although we would appreciate better feedback from some with respect to a vendor’s specific file format).
So please do understand that mentioning specific vendors has nothing to do with any positive or negative endorsements for said vendor. We’re merely stating facts with respect to the way how their techies have long time ago decided to organize their (giga)pixels.
Single or many files
As you already know from the previous article, virtual slides can consist of many different files. Within the files that represent MRXS slides, you find plenty of .dat files, for VSI slides you find .ets files (amongst others) etc.
Several vendors have adopted a single file format for their WSI data. These include Hamamatsu (NDPI), Aperio (SVS), Leica (SCN), and Zeiss (CZI and ZVI).
Other vendors that have adopted a multi-file approach include 3DHistech (MRXS), Olympus (VSI), and Motic (MDS).
The organization of data across multiple files for different vendors is not standardized. In the case of 3DHistech, individual files more or less represent different magnifications. Olympus’ file structure seems more organized around scanned regions of interest.
The single file formats can also be upgraded to multi-file formats. Hamamatsu scanners can create .ndpis files for fluorescent or z-stacked content. The “s” stands for “set”: the .ndpis file merely contains pointers to individual .ndpi files which then each contain the image for the particular layer or channel. You can open these .ndpi files by themselves by the way, but they’re only useful when you also correctly interpret their context from the .ndpis file.
Aperio SVS files can be accompanied by .xml files, which contain annotations. Ventana BIF files can be accompanied by .tifp and .bmp files.
As soon as you have more than 1 file involved, it becomes a multi-file file-format. The strict distinction is important, because some storage systems don’t support multi-file containers.
Examples
Let’s look at the 3DHistech line of scanners: In the screenshot below we scanned two slides: HE and PDL1. The software ends created “HE.mrxs” and “PDL1.mrxs” files, along with “HE” and “PDL1” subfolders.
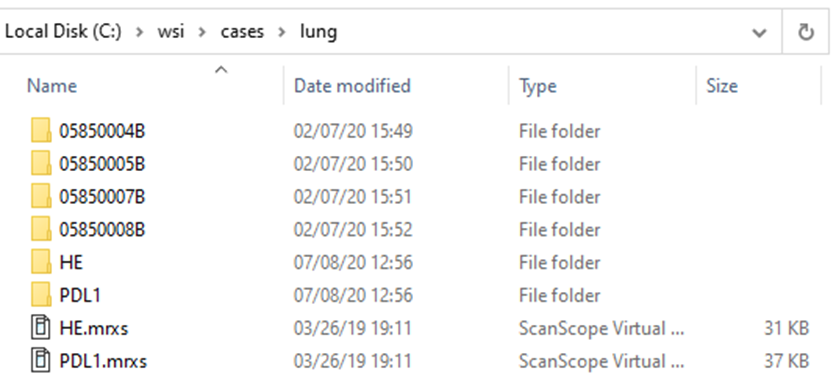
Within the subfolders, a standard naming convention it used, so you won’t find any more “HE*” files in there.
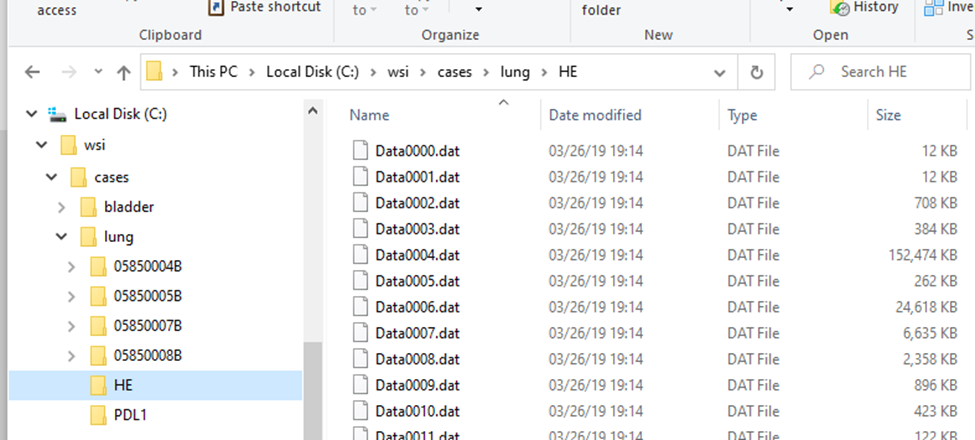
The .mrxs files themselves are typically just there to allow third-parties to identify these different file-types. Case in point: the .mrxs file can be renamed into a .jpeg file, and then you can just view it as any other image (it’s a quick way for us, too, to display slide thumbnails).
Another example? You got it!
This is what the structure of a .VSI slide looks like:
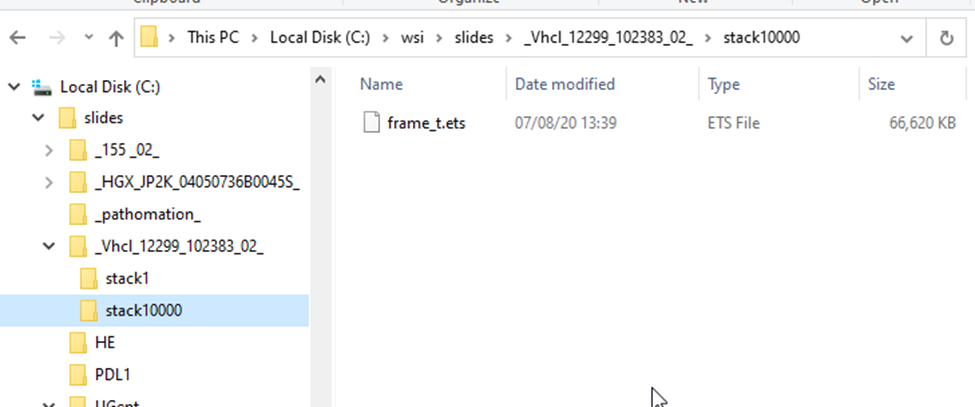
Each scanned region translates into a “stack”; each stack can contain one or multiple frames.
A different approach, a more hierarchical structure.
For DICOM slides, all related files are grouped together. No subdirectory required:
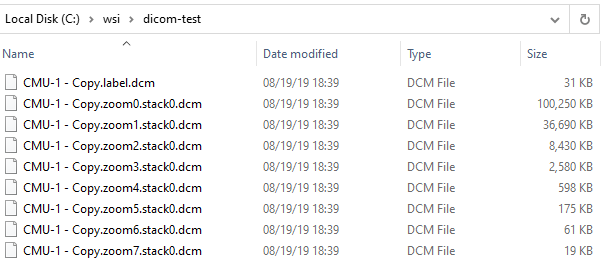
The details of these for the typical end-user don’t matter, except that they do matter when you want to copy / move/ transfer slides to others. The basic principle here to always make sure you not copy the index .vsi / .mrxs / .whatever file, but also all the accompanying data-files. Zipping them may help both you and your receiving party.
How Pathomation can help
Have a look at our [slides] folder in the Windows Explorer:
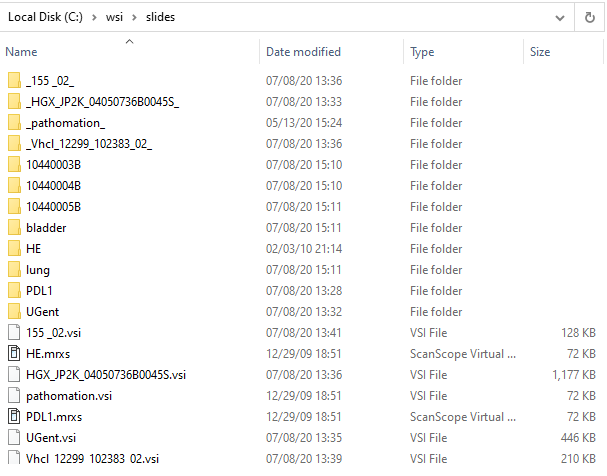
Confusing, right?
Now let’s have a look at the same folder through PMA.start eyes:
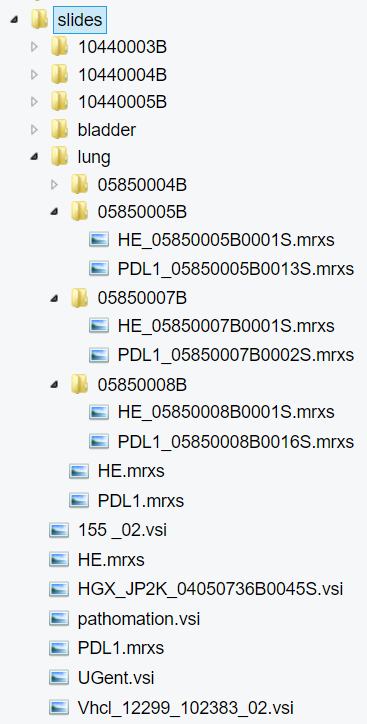
Since Pathomation knows slides, we systematically hide the intricacies of respective formats that you shouldn’t to worry about. In PMA.start it becomes obvious which subfolders are true subfolders, and which ones are merely there to support vendors’ data structures.
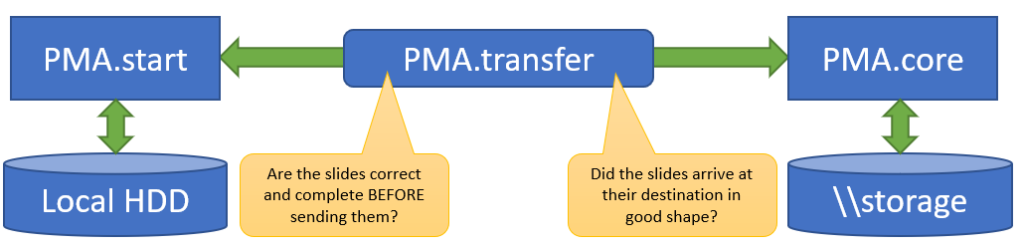
This doesn’t help you yet to transfer slides of course, but if you are one of our commercial users, you typically want to transfer slides from your local system to PMA.core and back. If that is you, then PMA.transfer is a great free tool (it’s part of your license package) for you to look at.
This is how our [slides] folder shows in PMA.transfer:
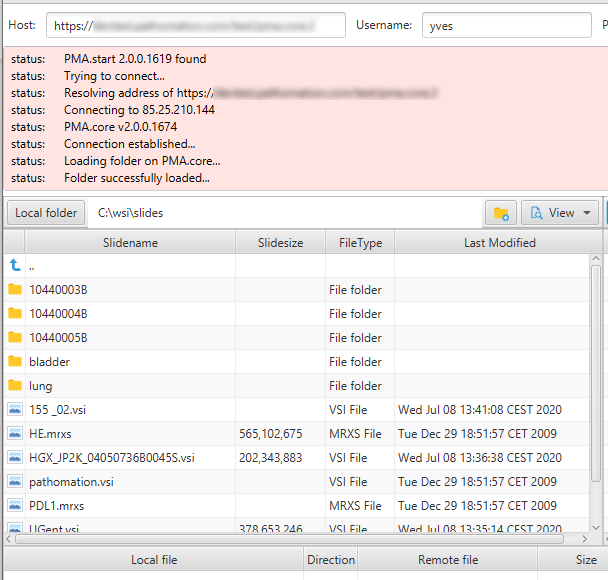
PMA.transfer seamlessly interfaces with PMA.start, and uses it as a jumping board to transfer slides between different endpoints. The biggest benefit of PMA.transfer therefore is that it encapsulates format-specific complexities and hides them from the end-user. Through PMA.transfer, you’re truly manipulating slides instead of files.
In addition, PMA.transfer also makes sure that only correct slides are transferred (nothing more frustrating than transferring 2 GB of data over Wifi, only to discover that the source was corrupt), as well as confirming that the transfer was completed successfully (probably the second most common source of frustration in the endeavors).
Why it matters
At Pathomation, we take much care designing our components in such a way that data duplication can be avoided at all costs. In PMA.control e.g., you can create as many cases and case collections as you want, but the data always remain in the same location. In PMA.core, you can create nested root-directories, each with different ACL properties. To your end-users these end up looking like different filesystems, but they’re really not. At the API level, we provide the possibility to fingerprint a slide, so you can scan for duplicate files and possibly eliminate them. All of these measures matter when you’re talking about Terabytes of data.
But there comes a time when you do need to copy slides. Perhaps you’re moving them from one installation to another, or there’s a network upgrade, or you just want to ship off some slides to a colleague (My Pathomation can now do this for you, too).
Whatever the case, (virtual) slides will need to be moved around. By their nature, they are big. It helps to have some understanding about how they are structured at that point.
And now you know…
… everything about whole slide images, or, at least, almost everything.
Amongst other things, .mrxs-, .vsi-, and other files help companies like ourselves decide what kind of file format is being used. The alternative would be that we find a large number of subfolders, and we have to parse each and everyone of those folders in a variety of ways to try to “guess” what file format it belongs to (if this is even the case at all; a subfolder can still be a regular subfolder, containing no slides at all). This would be a tremendous drag on system performance.
Whether you deal with Pathomation or another vendor, we hope this article has helped you take a peek behind the curtain of what whole slide images are made of, and how to best work with them.
Of course, if you are using the Pathomation platform, you should have a look at PMA.transfer, a no-hassle tool we developed to facilitate slide transfers.
If you’re not yet a Pathomation customer (whaaaaaaaaaat??), you can contact us for a free no-obligation demonstration.
