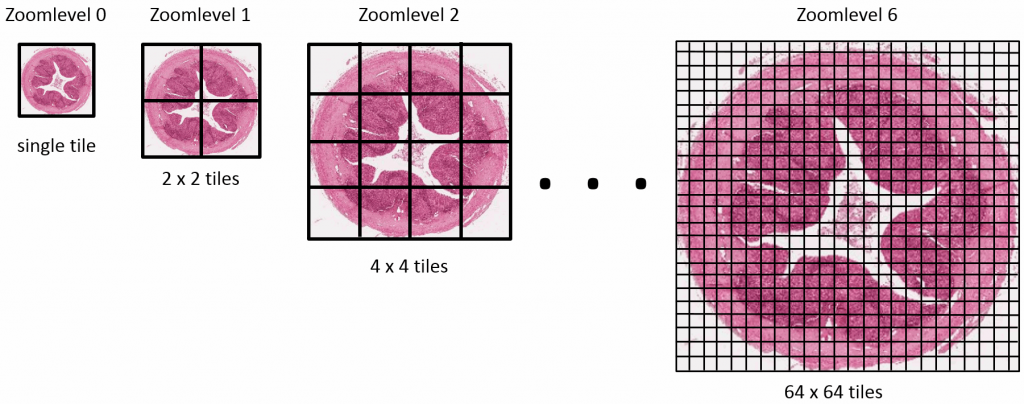
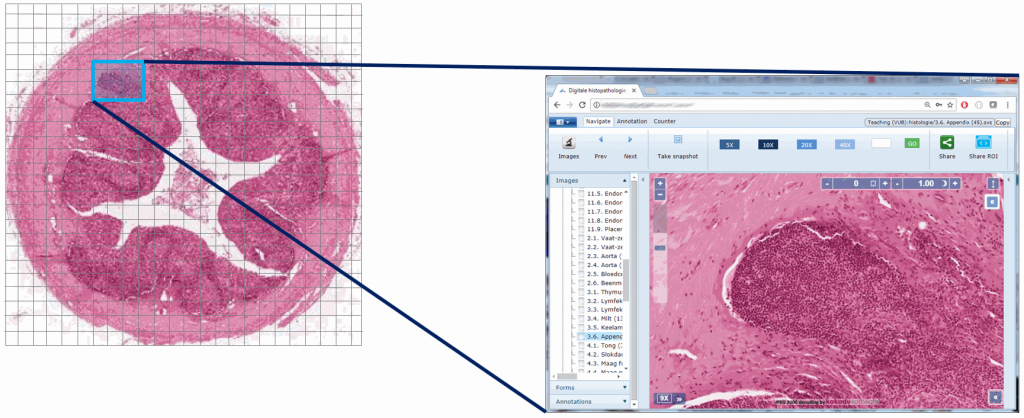
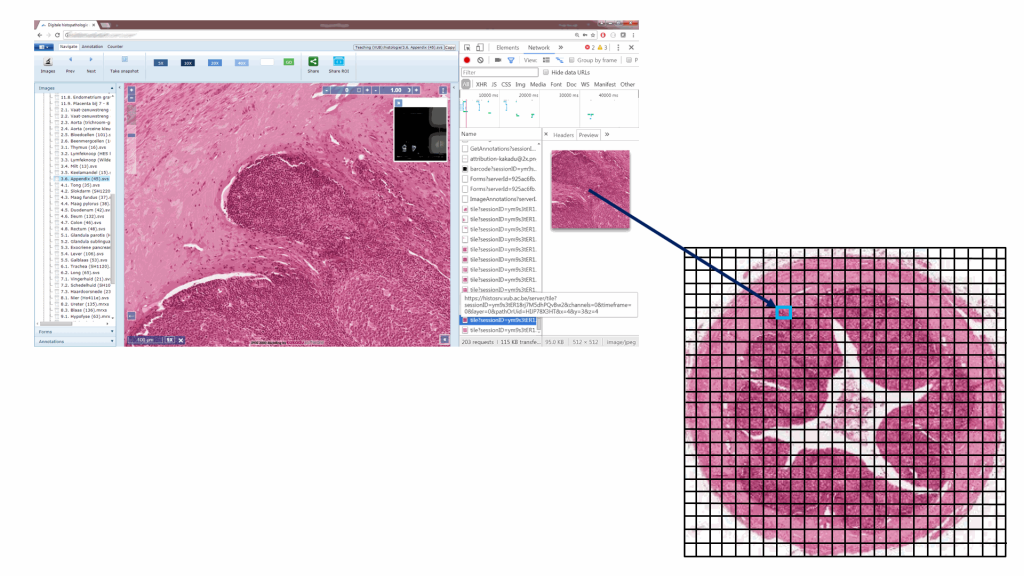
Standardization efforts in digital pathology
DICOM has been working on a standard description of digital pathology (DP) imaging data has been underway for a few years now. Digital pathology and whole slide imaging (WSI) is the focus of DICOM workgroup 26. A summary of its efforts can be found in David Clunie‘s paper in the Journal of Pathology informatics at https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6236926/
In 2014, we published our own conference paper on the effort (during the 12th European Conference on Digital Pathology in Paris, France) . The abstract is available through the Researchgate website; the full presentation from the ECP Paris conference is available through https://www.slideshare.net/YvesSucaet/digital-pathology-information-web-services-dpiws-convergence-in-digital-pathology-data-sharing
The focus of this blog is on imaging. To be complete, readers interested in digital pathology standardization efforts, should also have a look at the IHE PaLM initiative. Additional resources can also be found on slide 45 of our SlideShare publication.
Pathomation supports DICOM
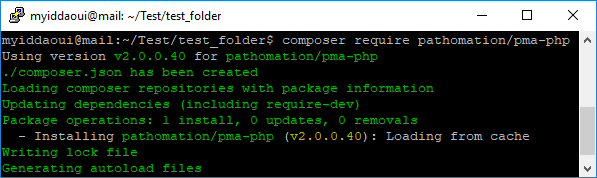
At Pathomation, we’ve been supporting DICOM supplement 145 file format extension (PDF available here) for a while now. We recently added our own “dicomizer” tool to our free PMA.start software. This is a command-line tool (CLI) that allows for the conversion of any WSI file format into a into a DICOM VL (Visible Light) Whole Slide Image IOD (Information Object Definition).
The Pathomation dicomizer is currently available on Windows only (other platforms pending) and can be installed as part of the regular setup process.
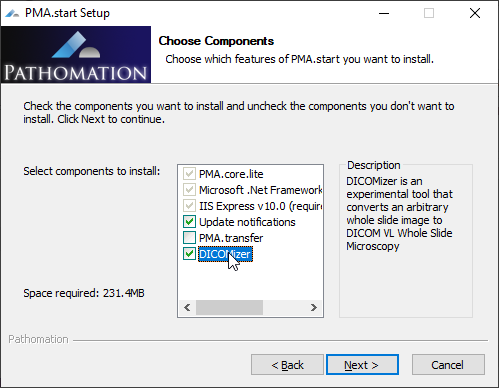
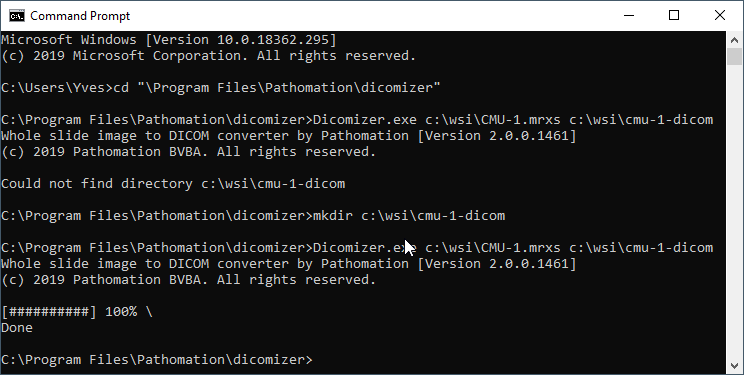
After installation, you have to navigate to the folder in which you installed the tool (typically c:\program files\pathomation\dicomizer) , and then you can invoke the tool through the command-line, like this:
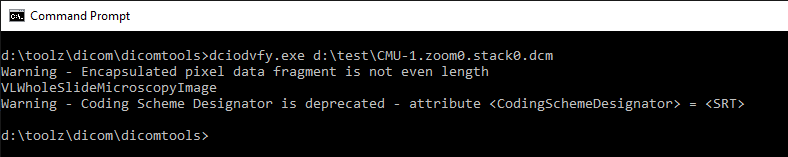
Running the validation tool from David Clunie on our generated DICOM slides results in the following:
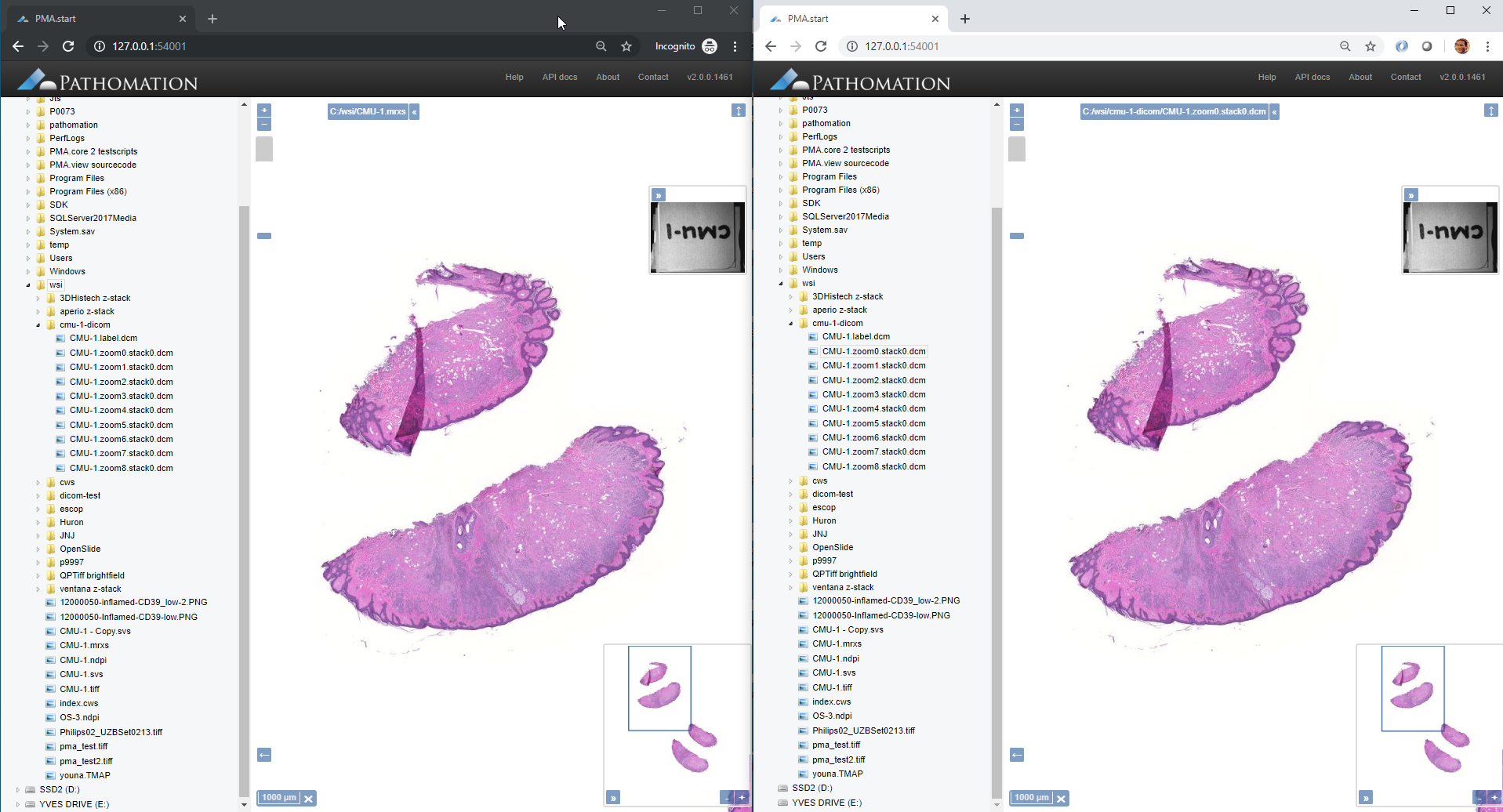
We can also visualize the slides side by side in two browser windows, empirically “proving” that the DICOM output is equivalent to the original slide:
So there you have it. DICOM has been involved in the digital pathology standardization process for a while now. For those interested to support it, you can now use Pathomation’s free dicomizer tool to get hands-on experience.