A naïve way to detect duplicate data
We often copy data for a variety of reasons. While testing a new program, we can copy data any number of times so the software is able to work on a dataset instead of a single datapoint. Temporary data unfortunately is all too often forgotten about, lingers around, and unnecessarily clutters our hard disks.
There are a number of ways to solve this problem. With Python, we can create a script that retrieves all slides, inspects the file size of each slide, and reports which two slides have the same size.
Using our PMA.python SDK, the code looks like this:
from pma_python import core
core.connect()
all_slides = core.get_slides("C:/", recursive = True)
def get_slide_size(slide):
info = core.get_slide_info(slide)
return info["PhysicalSize"]
all_sizes = {}
for slide in all_slides:
fp = get_slide_size(slide)
if "d" in fp.keys():
fp = fp["d"]
# print (slide, fp)
if not fp in all_sizes.keys():
all_sizes[fp] = []
all_sizes[fp].append(slide)
for (k, v) in all_sizes.items():
if len(v) > 1:
fn = v[0]
if not (".png" in fn.lower() or ".jpg" in fn.lower()):
print(k, len(v))
print(v)
But wait, what if two slides are the same size? Whole slide images are typically 100s of megabytes in size. Based on this characteristic, you could assume that it’s unlikely two slides result in identical file sizes. But macroscopic images are important in pathology, too, and then we’re talking about file sizes that are only a couple of megabytes in size at most. Now think of a scenario where tens of 1000s of cases are treated annually, with multiple macroscopic photos being taken of each resection piece… Suddenly the chances of two files having the same size becomes plausible.
There’s a second, albeit somewhat more hypothetical reason, why slide size is a poor indicator here. Wsi data could be stored in a container format, and the container format can have certain limitations. We observed e.g. that our PMA.start installation package has now not changed in size for the last 7 releases or so. But of course our code did change. So, empirically, file size is not a good discriminant for executable files. We feel therefore that we cannot assume that this would be the case for image file formats. Since re-scans are a specific concern with microscopy and WSI data, something better is needed than just the filesize.
Introducing fingerprinting
We can think of a way to unambiguously distinguish slides from one another by combining a number of characteristics into a digital fingerprint. These would include:
- Filesize (we didn’t say this was a bad one; just an insufficient one)
- Pixel size
- Pixels per micron
- Number of channels
- Number of z-stack layers
If we had infinite real-time computing power, we can think of more:
- Histogram https://en.wikipedia.org/wiki/Color_histogram
- A CRC32 checksum of the slide’s thumbnail https://en.wikipedia.org/wiki/Cyclic_redundancy_check
- An SHA-2 checksum on all physical slide content https://www.php.net/manual/en/function.hash.php
For practically, we define a slide’s fingerprint in the Pathomation platform as a combined hash https://en.wikipedia.org/wiki/Hash_function of physical file size, as well as most of the parameters returned through the GetImageInfo method.
We also consider this fingerprint method to be essential, and so for stability, it is incorporated at the level of PMA.core (PMA.start) itself rather than at SDK level, so it can be transferred across programming boundaries. A fingerprint for slide [foo] requested through PMA.java yields the same results as when requested through PMA.php or PMA.python.
Slide integrity
The fingerprint method is a good way to confirm the integrity of a slide itself. When a file is not what it pretends to be, the fingerprint cannot be calculated, and an error follows.

Note that the above would not be possible if we stuck to conventional CRC-like checks, since those don’t take into account the nature of a slide. Of course, you can do a CRC check on any file regardless of whether it actually is a slide or not.
Applications
We recently introduced PMA.transfer. Have you ever been frustrated by people sending you individual VSI or MRXS slides without anything else? Did you ever feel uneasy about just having transferred a gazillion number of bytes half across the world, without any reassurance of whether it actually worked? Then you should definitely have a look at PMA.transfer. It’s like FileZilla, but for slides. SlideZilla.
PMA.transfer uses fingerprinting to ensure data integrity in between transfers. Whether you’re moving slides from and to PMA.start, PMA.core, or My Pathomation, the same fingerprint calculation algorithm is used to compute a slide’s unique signature. This means that PMA.transfer can obtain the fingerprint of a source and a target instance and simply compare one with another to see that they’re identical.
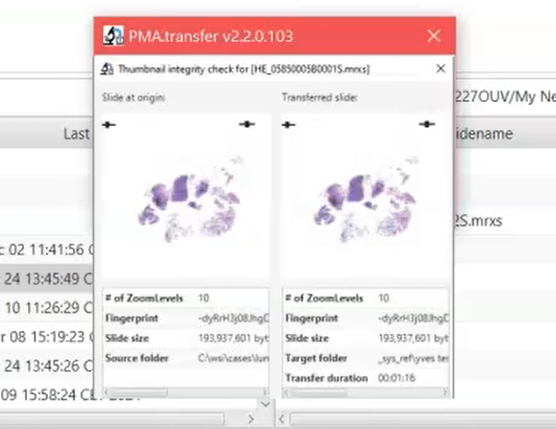
Another application is found in our upcoming PMA.studio product. Of course, the actual fingerprint of a slide is shown in the slide info panel. But a string like “” is not saying a whole lot and is for purely informative purposes at best.
PMA.studio can also be used to create annotations on slides. When you make an annotation, it is stored in our back end with a reference to the original slide, as well as the slide’s fingerprint.
This serves two purposes:
- After moving a slide to a new folder path or even physical location, you can still retrieve its annotations.
- When you have two identical copies of slides, each annotated separately, you can use fingerprinting to combine the annotations from both in a single view.
The possibility to combine annotations from identical slides stored in different locations in a single view offers opportunities for blinded studies and validation exercises. Inter- and even intra-observer variability can be measured this way, too.
Retrieving annotations by fingerprint is not available by default; you need to invoke this with an explicit button in the ribbon. It’s a performance thing.
Last but not least, fingerprinted annotations can be used to keep track of annotations during migration processes. As your applications for digital pathology increase, you will occasionally restructure your folder structures, or perhaps move to an entire new storage device altogether.
Finding duplicates
Back to our original question: imagine that you’ve been managing a whole slide repository for a while, and as careful as you’ve been, you suspect that you now have ended up with a number of copies of a variety of slides in different locations. You know: you copy a slide to test something, pinky-promise yourself that you’ll remove the slide again afterwards, that for real you’re really not going to forget this time… and then… you forget about it.
Thanks to the fingerprinting method and a few lines of Python, it is easy to trace duplicates however.
Here’s the basic code to build a dictionary that has all the possible fingerprints as keys. Each entry then contains a list that specifies where the exact copies that share a particular fingerprint:
slides_by_fingerprint = {}
for slide in slides:
fp = core.get_fingerprint(slide)
if not fp in slides_by_fingerprint:
slides_by_fingerprint[fp] = [slide]
else:
slides_by_fingerprint[fp].append(slide)
When a slide is unique, then the list of the dictionary will only have one entry. Alternatively, it can have two or more entries. So the duplicated slides are detected and flagged as follows:
for fprint in slides_by_fingerprint:
if len(slides_by_fingerprint[fprint]) > 1:
print(slides_by_fingerprint[fprint][0], "is copied", len(slides_by_fingerprint[fprint]), "times")
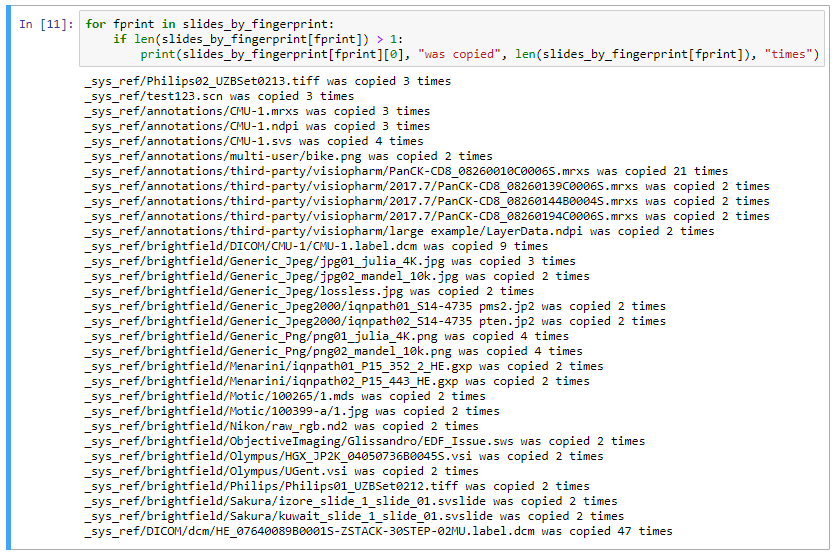
If you want, you can further automate the pruning and the deletion of these duplicates. Sometimes it’s easy; sometimes it’s not. You need to make sure that you have the original copy in its intended place. And in some cases, you may actually want to keep at least a second copy of a slide around, as one may be transient in a clinical setting, whereas its copy may have just been added to a reference repository to teach students and staff.
Coming full circle
Fingerprinting serves a triple function:
- Detect whether a dataset is a real slide or not
- Guard data integrity when transferring from one medium to another
- Trace slides and associated content through a complex storage hierarchy
Fingerprinting applies the concept of hash-functions to slides. Like everything in the Pathomation platform, the slide itself is the key unit to interact with. There is only one fingerprint for a slide, whether it consists of a single, or multiple files. Consequently, you can only obtain a fingerprint for a slide. If the file is somehow corrupt or the file format isn’t recognized by PMA.core, you’re not going to get a fingerprint from it. Last but not least, fingerprints are invariant across storage media and instances of PMA.core (PMA.start), making it a useful feature for slide tracking.
