The core
PMA.core is the centerpiece of the Pathomation software platform for digital microscopy. PMA.core is essentially a tile server. It does all the magic described in our article on whole slide images, and is optimized to serve you the correct field of view, any time, any place, on any device. PMA.core enables digital microscopy content when and where you want it.
Our free viewer, PMA.start, is built on top of PMA.core technology as well. PMA.start contains a stripped version of PMA.core, lovingly referred to as PMA.core.lite 🙂
PMA.core supports the same file formats as PMA.start does, and then some.
It acts as an “honest broker”, by offering pixels from as many vendor formats as possible.
Storage
With PMA.start, you’re limited to accessing slide content that’s stored on your local hard disk. External hard disks are supported as well, but at one point you end up with multiple people in your organization that need to access the same slide content. At that point, PMA.core’s central storage capabilities come into play.
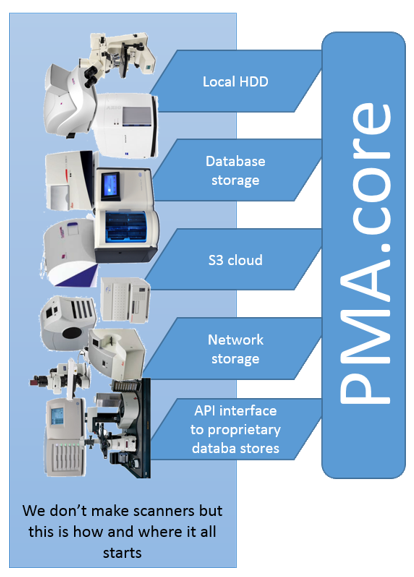
PMA.core is typically installed on a (Windows) server machine and can access a wider variety of storage media than PMA.start can. You can store your virtual slides on the server’s local hard disk, but as your data grows, this is probably not the place you want to keep them. So you can offload your slides to networked storage, or even S3 bucket repositories (object storage).
Pathomation does not, in contrast with other vendors, require a formal slide registration or import process to take place. Of course our software does need to know where the slides are. This is done by defining a “root-directory”, which is in its most generic terminology “a place where your slides are stored”.
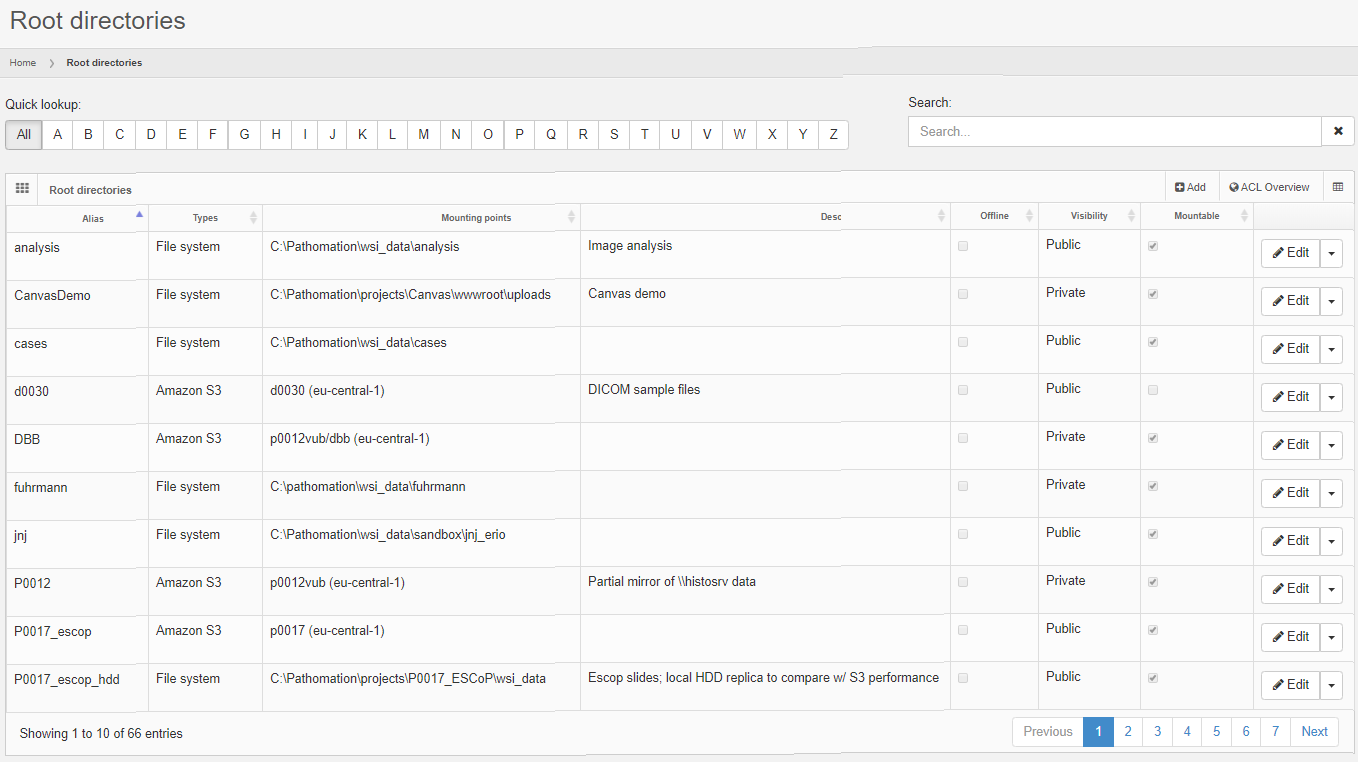
A root-directory can be location on the server’s hard disk, like c:\wsi. You can instruct your slide scanner to drop off new virtual slides on a network share, and likewise point PMA.core to \\central_server\incoming_slides\. Finally, you can store long-term reference material in an AWS bucket and define a root-directory that points to the bucket. The below screenshot shows a mixture of S3- and HDD-derived rootdirectories in one of our installations:
After defining your root-directory, the slides are there, or they are not, and representation of them is instant. An implication of this that you can manage your slides with the tools that you prefer; any way you want to. You can use the Windows explorer, or even using the command-line, should that end up being more convenient for you. Your S3 data can be managed through the AWS console, CloudBerry tools, or S3 explorer.
Security – Authentication
Another important aspect of PMA.core is access control. PMA.start is “always on”; no security credentials are checked when connecting to it. PMA.core in contrast requires authentication, either interactively through a login dialog, or automatically through the back-end API. In either case, upon success, a SessionID is generated that is used to track a user’s activity from thereon.
User accounts can be created interactively through the PMA.core user interface, or controlled through use of the API. Depending on your environment, a number of password restrictions can be applied. Integration with LDAP providers is also possible.
User accounts can be re-used simultaneously in multiple applications. You can be logged in through the PMA.core user interface, and at the same time use the same credentials to run an interactive script in Jupyter (using the PMA.core interface to monitor progress).
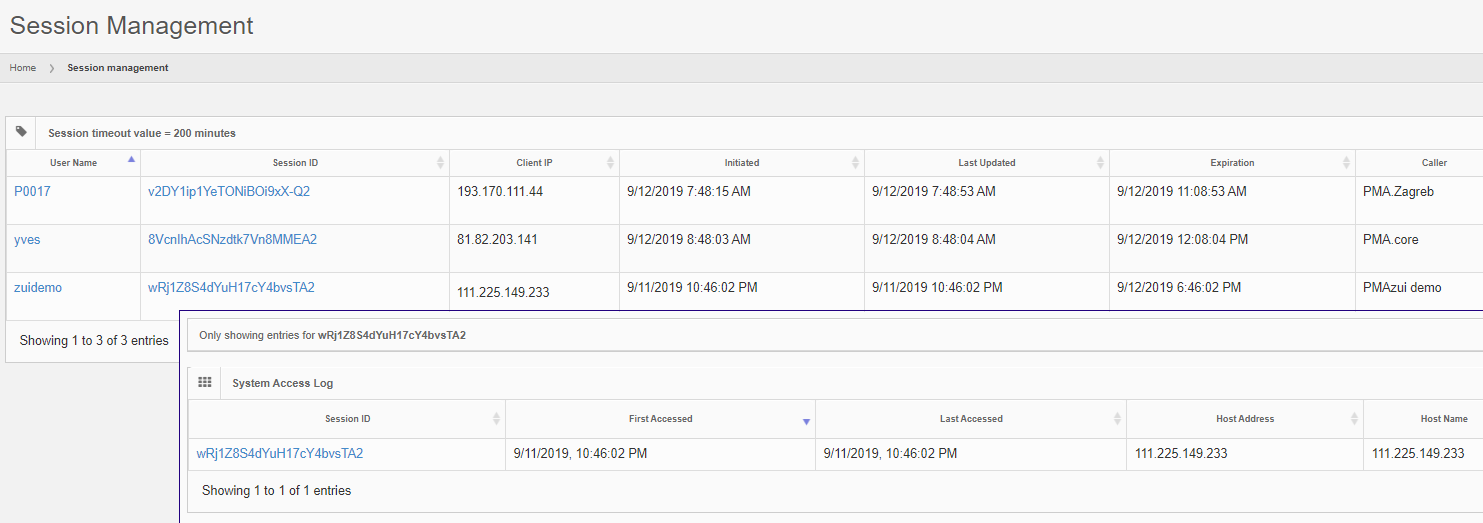
The interface in PMA.core itself at all times gives an overview what users are connected through what applications, and even allows an administrator to terminate specific sessions.
Security – Authorization
Our software supports authorization on top of authentication.
User permissions in PMA.core are kept simple and straightforward: a user account can have the Administrative flag checked or not, meaning that they can get access to PMA.core directly, or only indirectly through other downstream client application like PMA.view, PMA.control or the API. Another useful attribute to be aware of is CanAnnotate, which is used to control whether somebody can make annotations on top of a slide or not. Finally, an account can be suspended. This can be temporary, or can be mandated from a regulatory point of view as an alternative for deletion.
A root-directory can be tagged either as “public” or “private”. A public root-directory is a root-directory that is available to all authenticated users. In contrast, when tagged as “private”, the root-directory has an accompanying Access Control List (ACL) that determines who can access content in the root-directory.
The screenshot below shows the Administrative and Suspended flags for my individual user account, as well as what public and private root-directories I do or do not have access to:
Future versions of PMA.core can be expected to offer CRUD granularity.
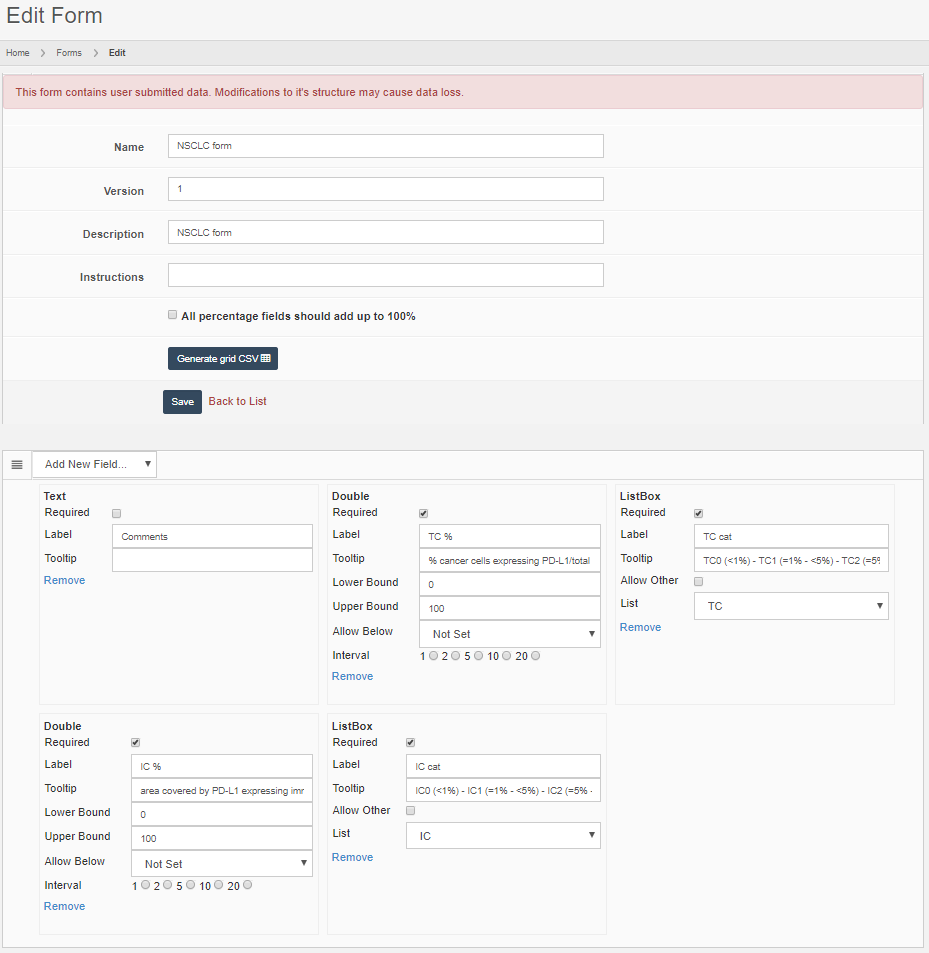
A powerful forms engine
Form data exists everywhere. Information can be captured informally, like the stain used, or as detailed as an Electronic Lab Request Form (ELRF). This is why Pathomation offers the possibility to define forms as structured and controllable data entities. A form can consist of a couple of simple text-fields, or be linked to pre-defined (ontology-derived) dictionaries. Various other Pathomation software platform components help in populating these forms, including PMA.view.
Forms can be accompanied by ACLs. In order to avoid redundancy, a form ACL consists of a list of root-directories rather then user accounts. In a project-oriented environment, it makes more sense that certain forms apply to certain root-directories which represent types of slides. Similarly, in a clinical environment, it makes sense to have slides organized in root-directories per application-type or by processing-stage. Freshly scanned slides that haven’t undergone a QA-check yet can be expected to have different form-data associated with them than FISH-slides.
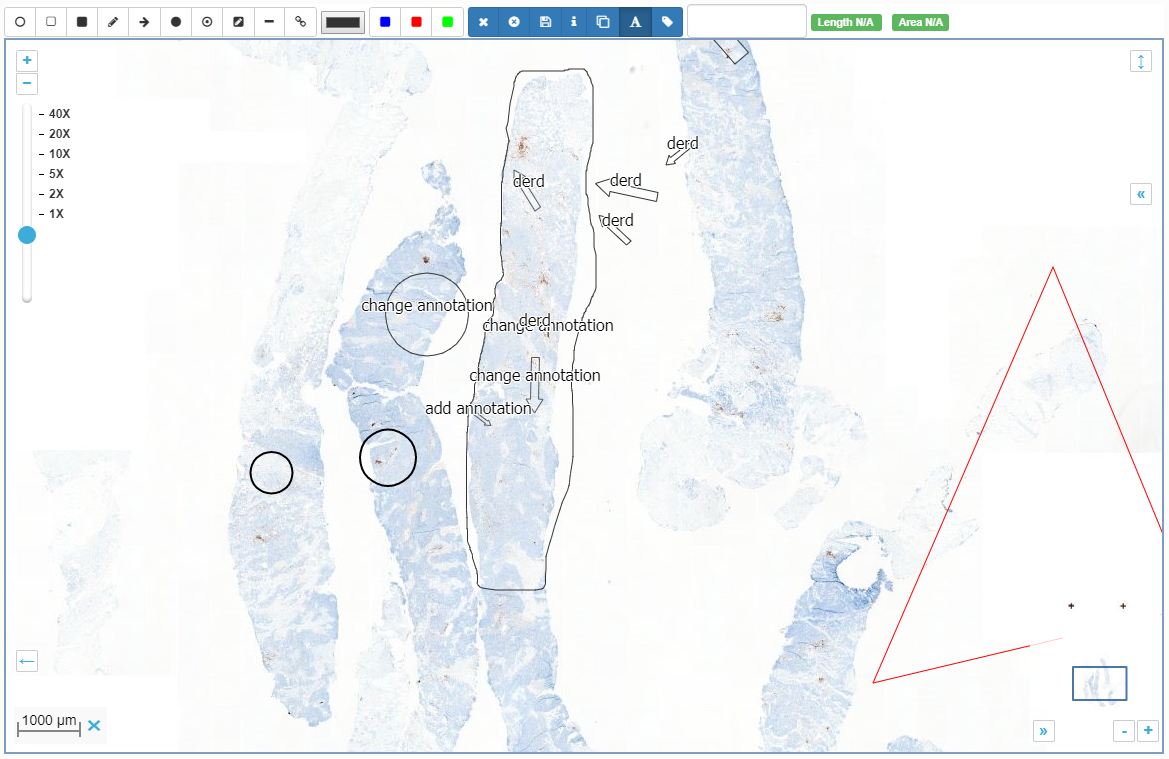
On-slide annotations
PMA.core support graphical on-side annotations. We support three types:
- Native annotations embedded within a vendor’s file format
- Third-party annotations coming from non-specific (image analysis) software
- Pathomation annotations
Pathomation-created annotations are the easiest to understand. You have a slide, and you want to indicate a region of interest on it. This region of interest can be necrotic tissue, or proliferated tumor cells. For teaching purposes, you could have a blood smear and highlight the different immune-celltypes.
Pathomation annotations are stored as WKT and can be anything that can be encoded in WKT (which is a lot). You need a downstream client to create them, but the basic viewer included in PMA.core can be used to visualize them, and our PMA.UI JavaScript framework can be used to create your own annotation workflows.
You could run an algorithm that does tissue detection and pre-annotates these regions for you.
In addition to making your own annotations, Pathomation can be used to integrate annotations from other sources. Certain file formats like 3DHistech’s MRXS file format or Aperio’s SVS file format have the ability to incorporate annotations. If you have such slides, the embedded annotations should automatically show when viewing the slide using any Pathomation slide rendering engine.
Last but not least, we can integrate third-part annotations. Currently, we support three formats:
- Visiopharm .mld annotation files
- Indica Labs .annotation files
- Aperio .xml annotation files (also used by others like Definiens)
Third-party as well as native annotations are read-only; you cannot modify them using Pathomation software.
Even more slide metadata
What about other structured data?
We think our forms engine is pretty nifty, but we’re not as arrogant (or clueless) to pretend that we foresee everything you ever want to capture in any form, shape, or size. It is also quite possible that a slide meta-database already exists in your organization.
For those instances where existing data stores are available, we offer the possibility to link external content. Rather than importing data into PMA.core (also a possibility actually), we allow you to specify an arbitrary connection string that points to an external resource that may represent an Oracle database. Your next step is to define the query to run against this resource, along with a field identifier (which can be a regular expression) that is capable to match specific records with individual slides.
Examples of external data sources can be:
- Legacy IMS data repositories that are too cumbersome to migrate
- Proprietary database systems developed as complement to lab experiments
- Back-end LIMS/VNA/PACS databases that support other workflows in your organization
Do try this at home
In this post, we’ve highlighted the main features of our PMA.core “honest broker” WSI engine aka tile server aka pixel extractor aka Image Management Server (IMS).
Warning: sales pitch talk following below…
If you’ve liked interaction with PMA.start and work in an organization where slides are shared with various stakeholders, you should consider getting a central PMA.core server as well. PMA.core is the center-piece of the Pathomation software platform for digital microscopy, and whether you prefer all-inclusive out-of-the-box viewing software, or are developing your own integrated processing pipelines, PMA.core can be the ideal middleware that you’ve been looking for. Contact us today for a demo or sandboxed environment where you can try out our components for yourself.
Ok, we’re done. Seriously, PMA.core is cool. Let us help you in your quest for vendor-agnostic digital pathology solutions, and (amongst others) never worry about proprietary file formats again.