Transferring whole slide images
The following is a cartoon from XKCD dating from a few years back. If you’ve ever been involved in the transfer of whole slide images, this will be very recognizable.
With whole slide imaging data, the above actually is more complicated. The step that is being skipped in the cartoon is the file integrity check. And with whole slide images, as a single virtual slide can consist of multiple files, it can become really complex really fast.
If you’re a professional user of PMA.core, you no doubt have already appreciated the convenience of the “integrity check” tool: it allows you to select a set of slides, and check if they actually really whole slide imaging data.
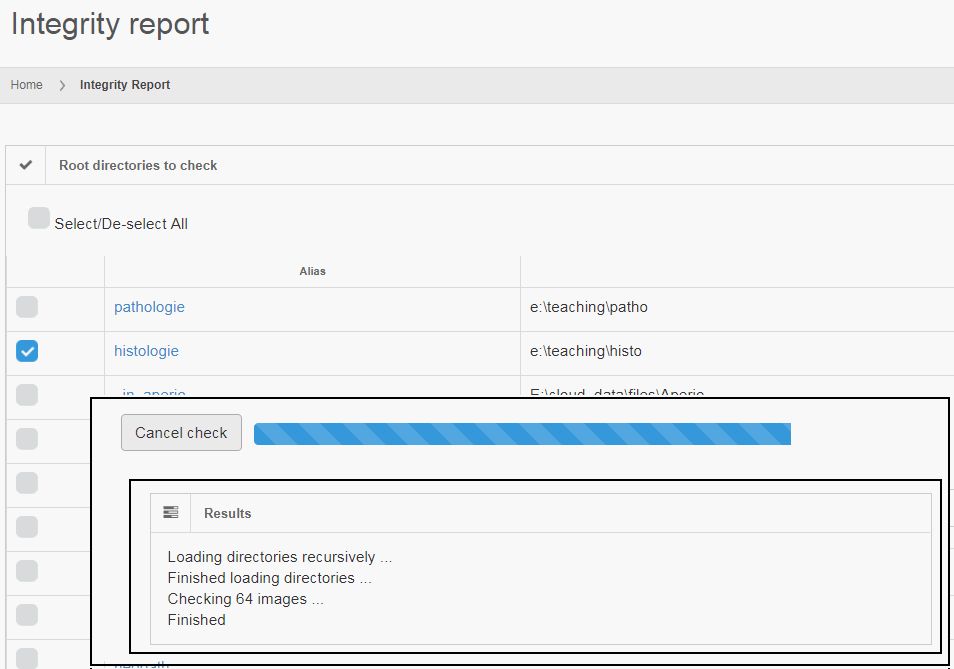
Can your image management solution do this? 🙂
But we admit, buying PMA.core just to verify virtual slide integrity is probably overkill.
DIY slide integrity
It turns out, PMA.start is just a well capable to verify virtual slide integrity. But first some background about why you would even want to do this.
The fundamental reason why you want to route all of your slide manipulation operations through PMA.start or its bigger brother PMA.core is that the software knows and understands what a slide is. When you copy a number a multi-file slides from one device to another and you refer to our software components in your code for verification purposes, you cannot make any mistakes in terms of accidentally copying only parts of files the slides. If a mistake does crop up in your report, you can simultaneously check whether something was perhaps wrong with the original slides and files. Digital pathology is hard enough as it is; at least do it on a unit of data (the “slide” rather than the “file”) that makes sense!
As much of slide transfers occur “ad hoc” (on a “need to transfer” basis sort of speak), we wrote an integrity check procedure ourselves as an Anaconda Jupyter notebook.
In order to get PMA.python to work with Anaconda, you’ll first have to get it through PyPI. Start an anadonda prompt and type in “pip install pma_python”.
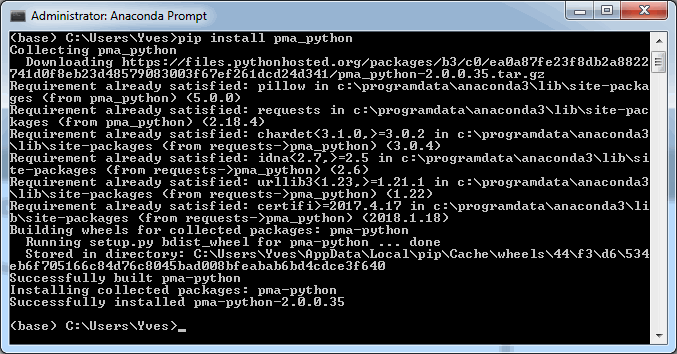
Great, now you’re ready to import our nifty library in your Jupyter notebooks.
Here’s a good way to start all of your notebooks:
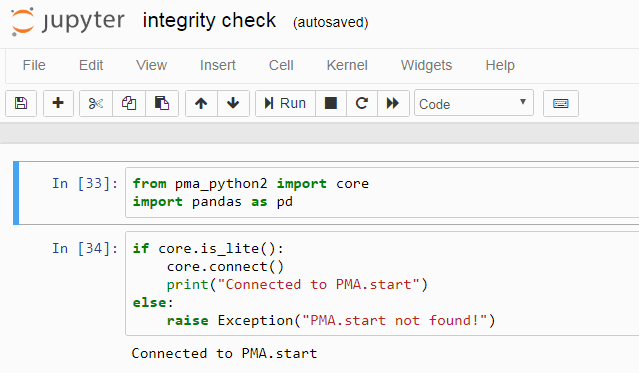
The first block confirms the availability of the library; the second block confirms that PMA.start is running on your system.
This is where Jupyter really shines: you can subdivide your Python (or R) code in separate discrete parts and execute them in an order you want. It becomes really convenient to tweak your scripts and even faster to debug them.
Our strategy for doing an integrity check on a folder an its subfolders is straightforward: loop over all the slides in the folder and see whether we can request the slide information from each one:
def loop_over_directories(dir):
slides = {}
for subdir in core.get_directories(dir):
slides.update(loop_over_directories(subdir))
for slide in core.get_slides(dir):
slides[slide] = core.get_slide_info(slide)
return slides
The result of this function is a dictionary that as keys has paths to all the slides, and as values has information objects (or None in case the slide turned out to be corrupt).
We can now invoke the function on an external hard disk’s folder after copying to see if it goes ok. By means of a sanity check, we’ll also output some parameters on the first slide encountered, as well as the number of items in the dictionary:
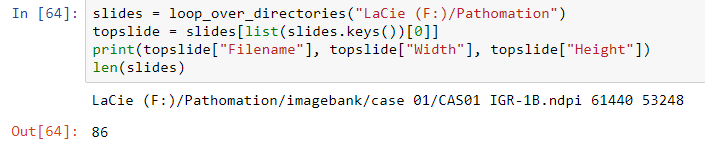
slides = loop_over_directories("LaCie (F:)/Pathomation")
topslide = slides[list(slides.keys())[0]]
print(topslide["Filename"], topslide["Width"], topslide["Height"])
len(slides)
The output in Jupiter looks like this:
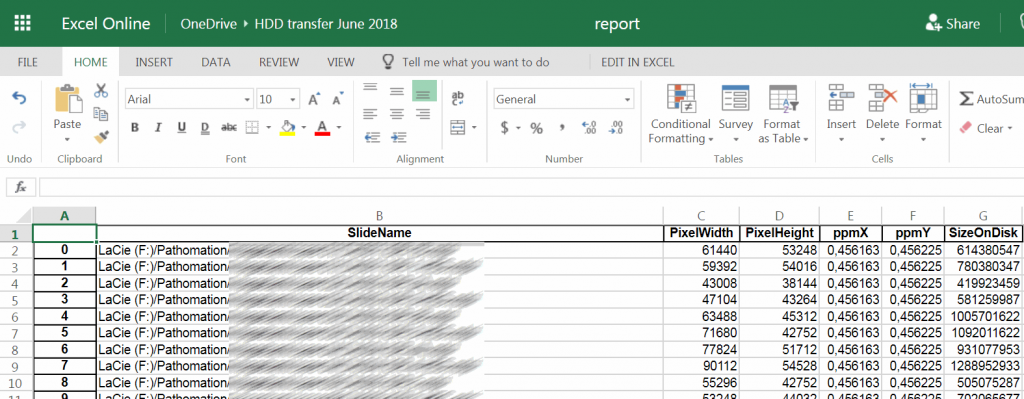
Our next step is to convert part of the dictionary data to a pandas DataFrame, which in turn it written out as an Excel file. The Excel file is subsequently stored on the external hard disk to provide a check-list for the client and confirm data integrity on their end upon receipt.
df = pd.DataFrame(columns=["SlideName", "PixelWidth", "PixelHeight", "ppmX", "ppmY", "SizeOnDisk"])
df.set_index(["SlideName"])
for slide in slides:
info = slides[slide]
if info == None:
print("Unable to get slide information for", slide)
else:
df = df.append({"SlideName": slide,
"PixelWidth": info["Width"],
"PixelHeight": info["Height"],
"ppmX": info["MicrometresPerPixelX"],
"ppmY": info["MicrometresPerPixelY"],
"SizeOnDisk": info["PhysicalSize"]}, ignore_index=True)
print (df.shape[0], " out of ", len(slides), " check out OK")
Great, but what if things don’t work out they’re supposed to?
As it turns out, we had been copying a collection of slides (about 60GB in total) overnight from an FTP site. Chances are that in the time that takes, something will go wrong. This turned out have happened in our case, as well, and the output flagged correctly that there was a mistake.
The flagged item was minor actually; one of the files had indeed failed to copy and had resulted in a file size with 0 bytes. We downloaded the file anew, and now everything checked out.
Did we need PMA.start to find out that a file was the wrong size? No, we could have used a range of other tools. But the point is that many things can go wrong with whole slide image transfer. The final goal is to check slide integrity, and once a problem is detected, other tools can be used to dig further and resolve them.
There are other potential problems, too: you may be sending someone slides who’s unable to read them. By including a test report from your end with the data, you’re telling them that the files are indeed all right, and that other problems may be at play (perhaps they’re not using the right viewer or import module for the file format that you sent them).
Troubleshooting and debugging is all about eliminating possibilities. We like to think PMA.start (and it’s bigger brother PMA.core) can help you get there faster than anything else on the market today.
You can download the Jupyter notebook discussed in this post. Just modify the folder_to_check variable in the third codeblock to point to the location that you want to validate.