This is a follow-up blog post. If you missed the first part, you can read it here: WSIs and pseudonymization.
De-identification
De-identification, is a term used for any process of reducing personal information, aiming at increasing personal privacy and data compliance. For example, pseudonymization is a de-identification procedure, further de-identification steps may lead to anonymization.

Pseudonymization
Essentially, pseudonymization consists in the replacement of direct identifiers by a pseudonym. It permits the WSI to be traced back to the patient as it is often needed in the clinical practice, while allowing it to be shared with other areas of medical research, often subject to different legal agreements. Due to its recognized importance, in practice all patient samples entering a pathology lab are pseudonymized at the lab registration step, each new sample gets a sequential biopsy number. In the specific case of research projects, pseudonymization is sufficient on the condition there is patient consent and approval of an Ethical Committee.
Anonymization
Anonymization, is the most radical de-identification action, – it ensures patient privacy. In the specific case of the WSI, it requires the erasure of any written data/metadata entry from the WSI so that the data is irreversibly altered in such a way that a data subject can no longer be identified directly or indirectly, either by the data controller alone or in collaboration with any other party (ENISA, 2019).
Additional security steps for pseudonymized data
Further measures can be employed to secure patient data from unauthorized access. Increasing the number of steps and gatekeepers needed to identify a patient, increases security. For example, Provenance information management, where a WSI can be traced back to the original biological material, without exposing patient information might be employed to further protect the likelihood of unauthorized access, despite the use of pseudonymization. (Holub et. al; 2022).
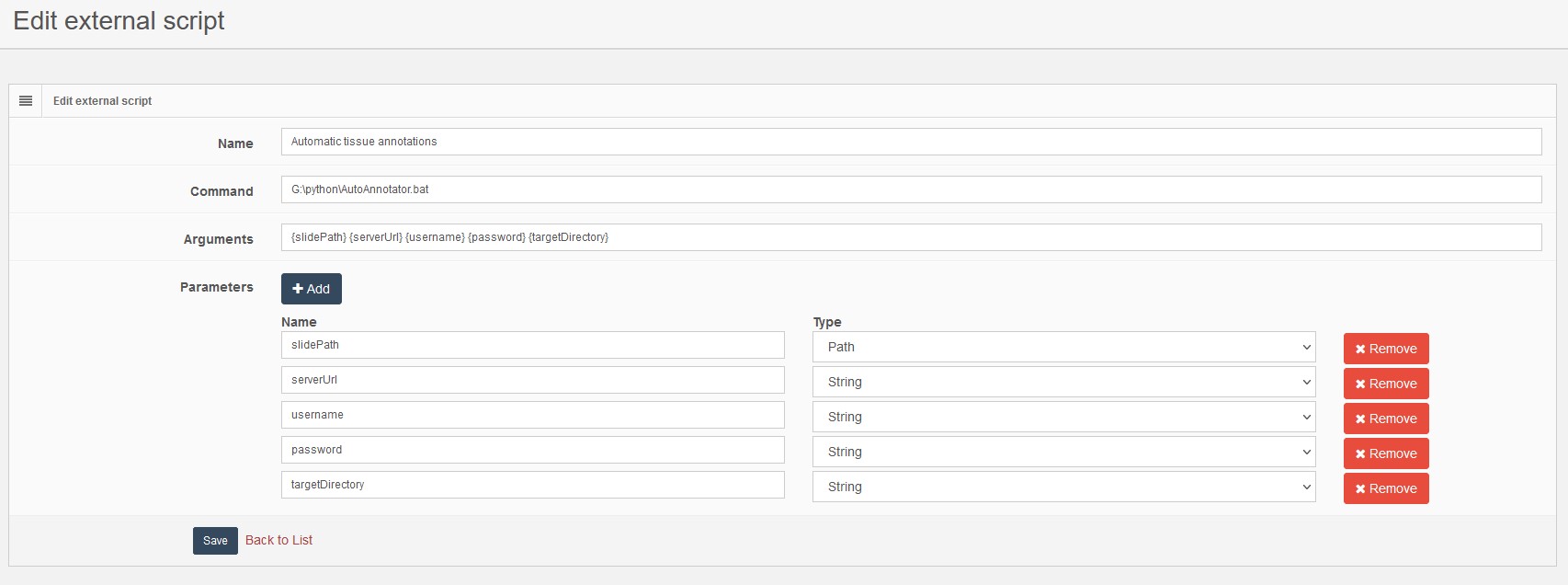
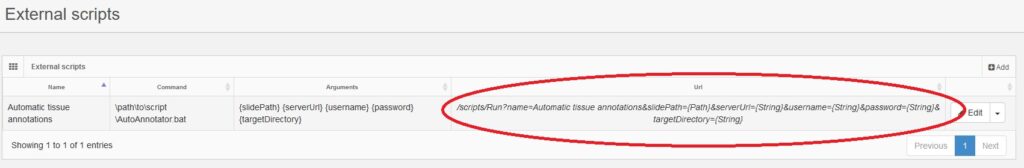
Pathomation’s PMA Core: a security and privacy gatekeeper
At Pathomation we offer a series of tools and configurable options to ensure data compliance with major personal data privacy laws (GDPR, HIPAA). As data processors, whenever we store client data in our servers, the data is received pseudonymized, – Scenario 3 of the ENISA guidelines “[…]the data controller [our clients] again performs the pseudonymisation but this time the processor is not involved in the process but only receives the pseudonymised data from the controller.”
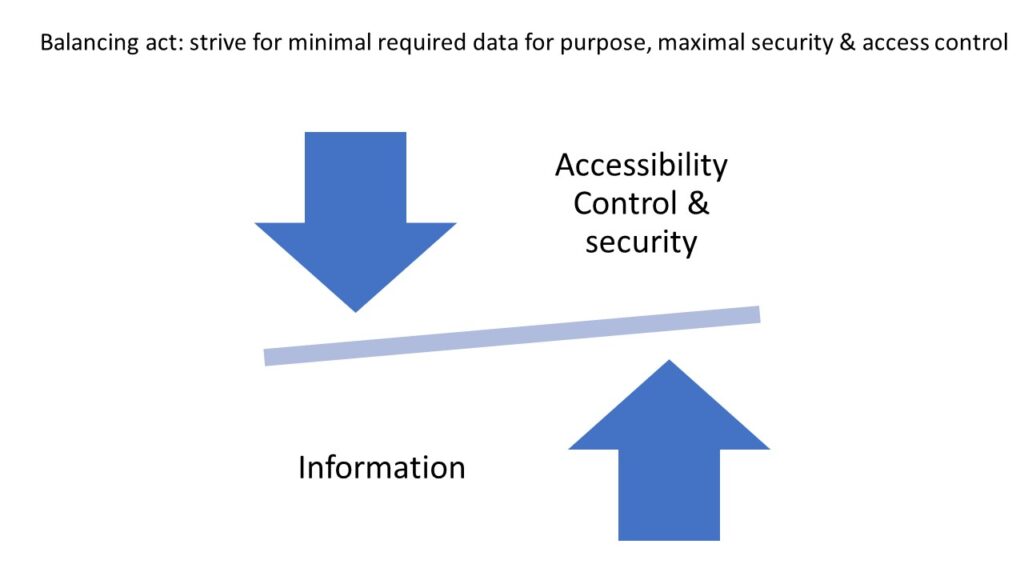
Despite managing pseudonymized data, which confers a considerable degree of privacy, the design of our IT infrastructure and systems, ensures that the data is kept secure and access is controlled.
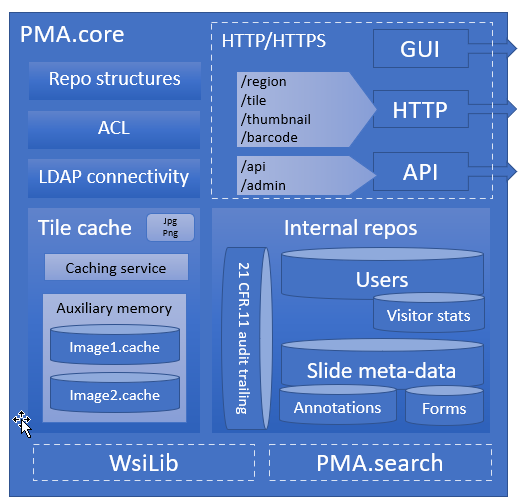
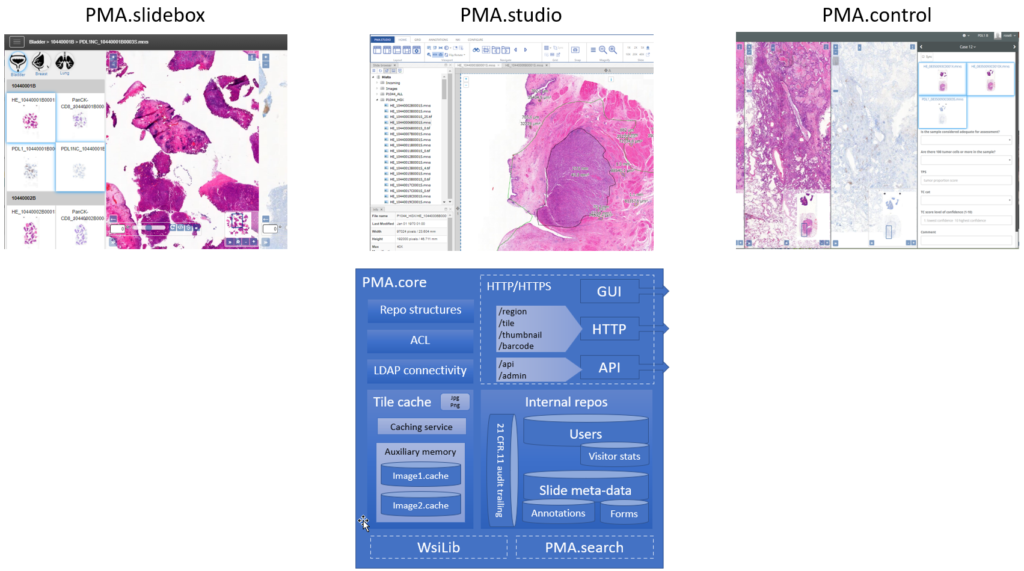
For example, our Image Management Solution (IMS) PMA.core, the centerpiece in our software platform for everything slide-management, is also the guardian of sensitive information. PMA-core is CE-IVD labelled for clinical diagnostic use, offering best practices and data management tools covering among others, robust permissions, encryption, passwords, audit trailing and data anonymization tools.
Thinking of data management practices aiming at ensuring data privacy in a daily workflow, with PMA-core is now possible to incorporate the following management practices and tools into your workflow:
De-identification tools: Beyond pseudonymization
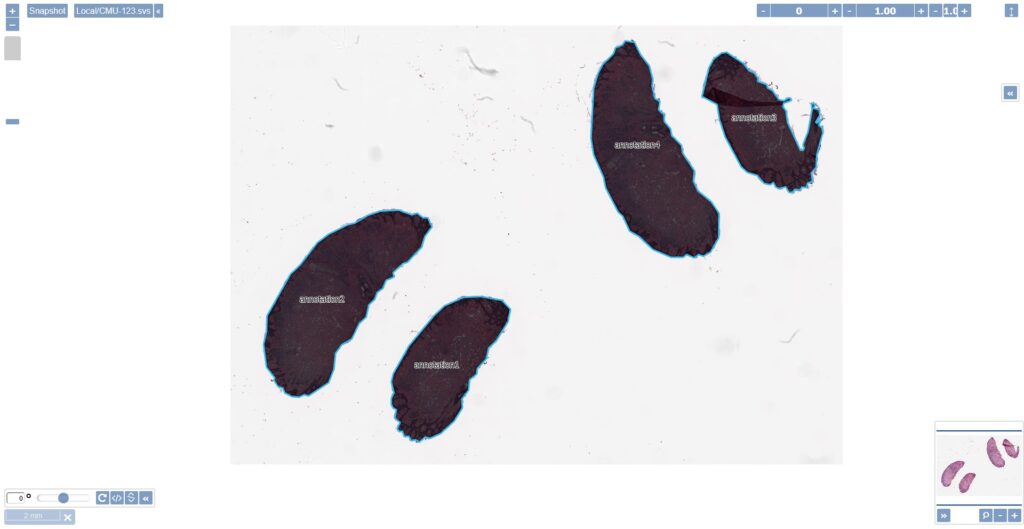
When presenting a viewport to serve slide content, one can opt to hide elements in the viewport that could lead to inadvertently revealing sensitive information: both the label (barcode) and filename widgets can easily be hidden with a single line of JavaScript code. Check our online viewport configuration demo for relevant technical information on this.
The viewport configuration is useful in an environment where sometimes you do, yet sometimes you don’t want to show slide identification information. At root-directory level, therefore, one can opt to stipulate that label information is never to be revealed. A hybrid environment can thus be created in a medical school, where human pathology samples for teaching can be prevented from ever exposing label information, while in diagnostic routine these remain visible.
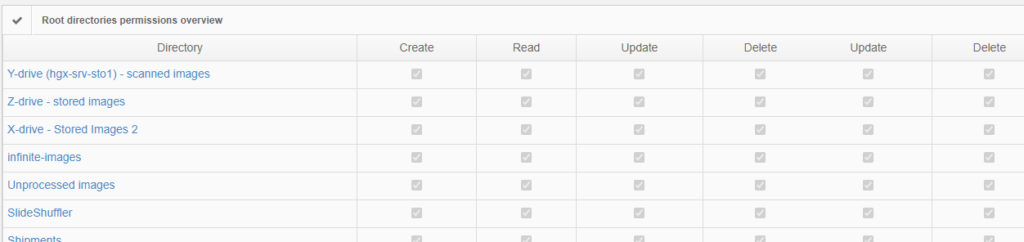
Permissions
The above are software solutions. PMA.core never manipulates the original raw data. This means that the original whole slide image will always retain the original slide label information unless you opt to anonymize the data. Hypothetically somebody could download an original slide from one PMA.core, and open it somewhere else. In order to prevent people from downloading selected data that they’re not allowed to see, PMA.core as a final back-stop supports granular permission settings: you can hence specify that people can view a slide (which essentially means piecemeal controlled serving of individual tiles), but they’re not allowed to download it; meaning they could never gain access to the original data.
Anonymization
Eventually you decide that a particular image or slide collection can (or must be) anonymized. In fact, whenever you need to share WSIs publicly such as along a congresses and other symposia or provide pathology training, any information that could lead to patient identification should be removed. In other words, pseudonymization is no longer enough, you must anonymize the WSI. In that case we offer you our in-house developed tool, Dicomizer. It is possible to export any WSI file format into a Dicom standard image, while at the same being able to remove even metadata embedded in the image files, together with the label on the WSI.
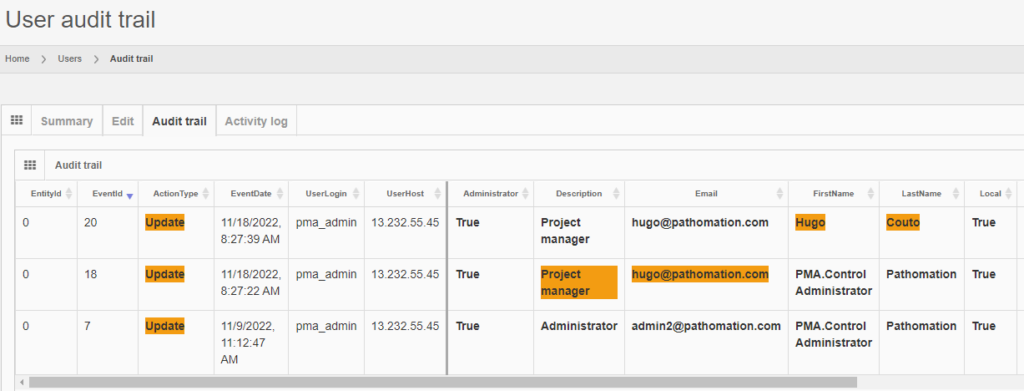
Trail Audit
Under GDPR, data processors have a legal obligation to maintain records of their processing activities. We facilitate this obligation by providing real time trail audit; which translates in the ability to understand who has used a particular image and when.
On top of all these, we love to bespoke our applications and services to accommodate your very specific needs.
Glossary of terms
- GDPR (ambit 26) The principles of data protection should apply to any information concerning an identified or identifiable natural person. Personal data which have undergone pseudonymisation, which could be attributed to a natural person by the use of additional information should be considered to be information on an identifiable natural person.
- Datacontroller/dataprocessor: Under GDPR, there are two key entities: the data controller determines the purposes for which and the means by which personal data is processed. The ‘why’ and ‘how”. The data processor processes personal data on behalf of the controller. The controller can be also the processor.
- Pseudonymization: GDPR (Article 4(3b) “the processing of personal data in such a way that the data can no longer be attributed to a specific data subject without the use of additional information, as long as such additional information is kept separately and subject to technical and organizational measures to ensure non-attribution to an identified or identifiable individual.”
- Anonymization: Recital 26, EU GDPR: “(…)information which does not relate to an identified or identifiable natural person or to personal data rendered anonymous in such a manner that the data subject is not or no longer identifiable.”