The legacy
When you say digital pathology or whole slide imaging, sooner or later you end up with OpenSlide. This is essentially a programming library. Back in 2012, a pathologist alerted me to it (there’s some irony to the fact that he found it before me, the bioinformatician). He didn’t know what to do with it, but it looked interesting to him.
OpenSlide is how Pathomation got started. We actually contributed to the project.
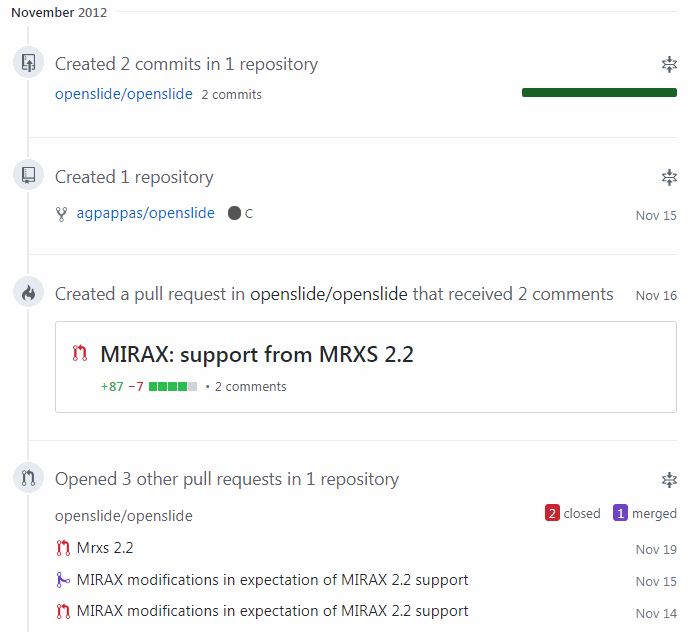
We also submitted sample files to add to their public repository (and today, we use this repository frequently ourselves, as do many others, I’m sure).
Our names are mentioned in the Acknowledgements section of the OpenSlide paper.

But today, the project has rather slowed down. This is easy to track through their GitHub activity page:
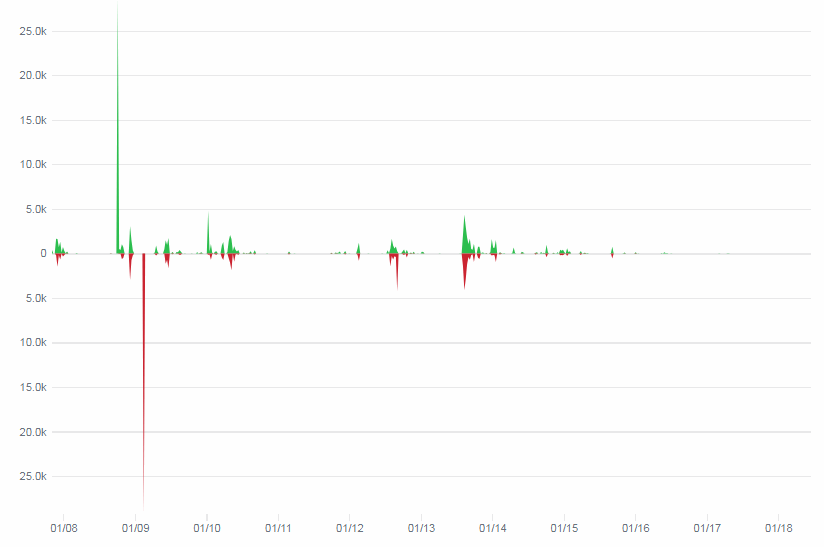
Kudos to the team; they do still make an effort to fix bugs that crop up, today’s but activities seem limited to maintenance. Of course there’s a possibility that nobody gets the code through GitHub anymore, but rather through one of the affiliate projects that they are embedded into.
Consider this though: OpenSlide Discussions about supporting support for (immuno)fluorescence and z-stacking date back to 2012 , but never resulted in anything. Similarly, there are probably about a dozen file formats out there that they don’t support (or flavors that they don’t support, like Sakura’s SVSlide format, which was recently redesigned). We made a table of the formats that we support, and they don’t, at http://free.pathomation.com/pma_start_omero_openslide/
Free at last
Our own software, we’re proud to say, is OpenSlide-free. PMA.start no longer ships with the OpenSlide binaries. The evolution our software has gone through then is as follows (the table doesn’t list all the formats we support; for a full list see http://free.pathomation.com/formats):
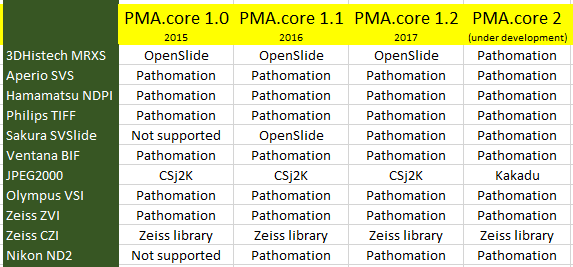
At one time, we realized there was really only one file format left that we still ran through OpenSlide, and with the move to cloud storage (more on that in a later post), we decided we might as well make one final effort to re-implement a parser for 3DHistech MRXS slides ourselves.
Profiling
Of course all of this moving away from OpenSlide is useless if we don’t measure up in terms of performance.
So here’s what we did: we downloaded a number of reference slides from the OpenSlide website. Then, we took our latest GAMP5 validated version of our PMA.core 1.2 software, and rerouted it’s slide parsing routines to OpenSlide. This result in a PMA.core 1.2 build that instead of just 2 (Sakura SVSlide and 3DHistech MRXS), now reads 5 different WSI file formats through OpenSlide: Sakura SVSlide, 3DHistech MRXS, Leica SCN, Aperio SVS, and Hamamatsu NDPI.
Our test methodology consist of the following steps:
For each slide:
Determine the deepest zoomlevel
From this zoomlevel select 200 random tiles
Retrieve each tile sequentially
Keep track of how long it takes to retrieve each tiles
We run this scenario of 3 instances of PMA.core:
- PMA.core 1.2 without OpenSlide (except for SVSlide and MRXS)
- PMA.core 1.2 custom-build with OpenSlide (for all 5 file formats)
- PMA.core 2 beta without OpenSlide
The random tiles selected from the respective slides are different each time. The tiles extracted from slide.ext on PMA.core instance 1 are different from the ones retrieved through PMA.core instance 2.
Results
As simple as the above script sounds, we discovered some oddities while running them for each file format.
Let’s start w/ the two easiest to explain ones: SVS and SVSlide.
A histogram of the recorded times for SVSlide yields the following chart:
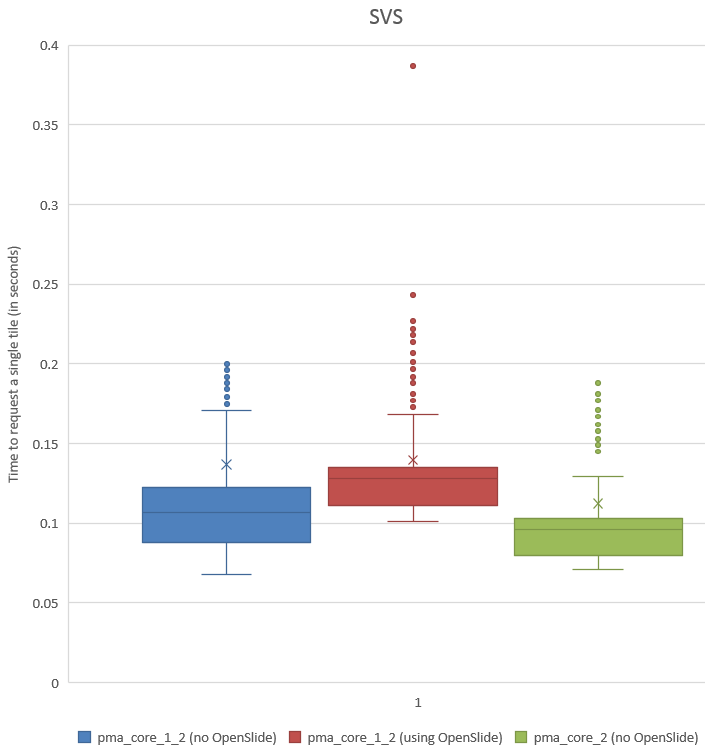
We can see visually that PMA.core 1.2 without OpenSlide is a little faster than the PMA.core 1.2 custom-build with OpenSlide, and that we were able to improve this performance yet again in PMA.core 2.
The p-value confirm this. First we do an F-test to determine what T-test we need (p-values of 1.66303E-78 and 2.03369E-05 suggests inequal variances),
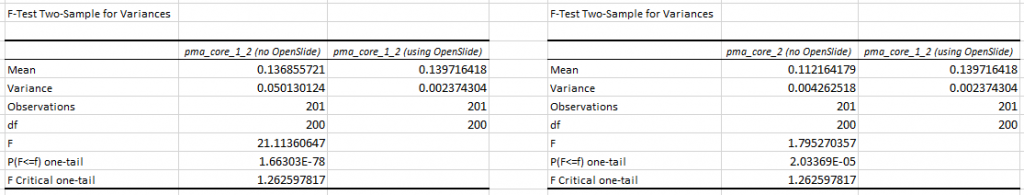
Next, we find a p-value of 0.859 (n=200) for the difference in mean tile retrieval time between PMA.core 1.2 with and without OpenSlide, and a p-value of 2.363E-06 for the difference in means between PMA.core 1.2 with OpenSlide and PMA.core 2 without OpenSlide.
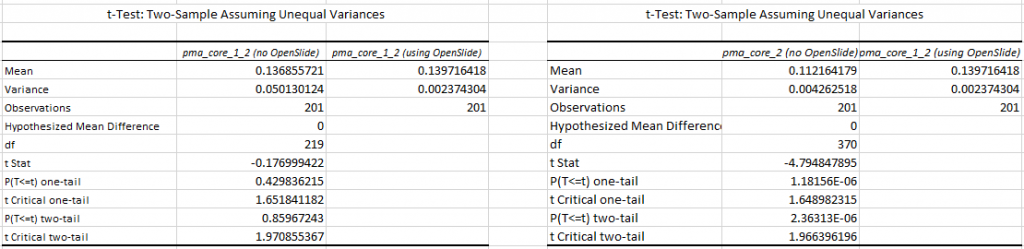
We see this as conclusive evidence that PMA.core 2 (as well as PMA.start, which contains a limited version of the PMA.core 2 code in the form of PMA.core.lite) can render tiles faster than OpenSlide can.
What about SVSlide?
Again, let’s start by looking at the histogram:
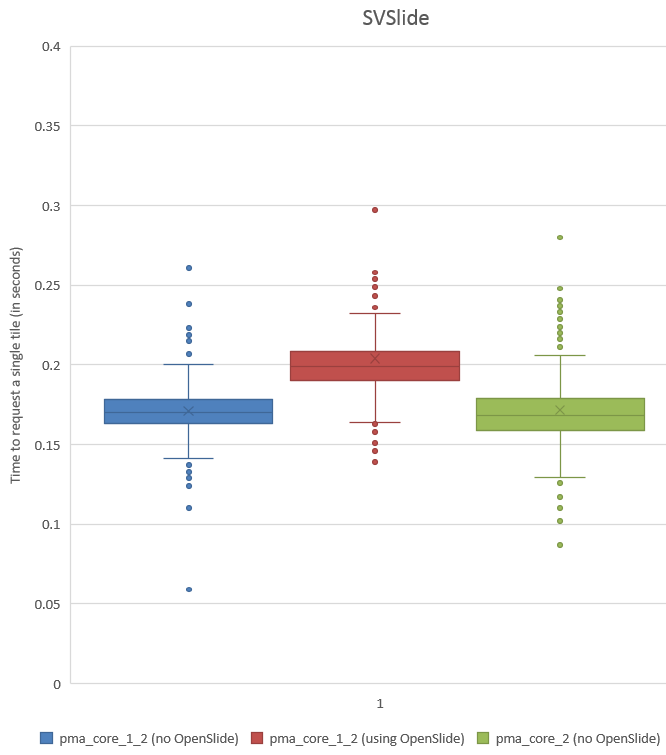
This is the same trend as we saw for SVS, so let’s see if we can confirm this with statistics. The F-statistic between PMA.core 1.2 with and without OpenSlide yields a p-value of 3.064E-22; between PMA.core 1.2 without OpenSlide and PMA.core 2 we get a p-value of 3.068E-13.
Subsequent T-tests (assuming unequal variance) between PMA.core 1.2 with and without OpenSlide show a p-value of 8.031E-19; between PMA.core 1.2 with OpenSlide and PMA.core 2 we get a p-value of 4.10521E-17 (one-tailed).
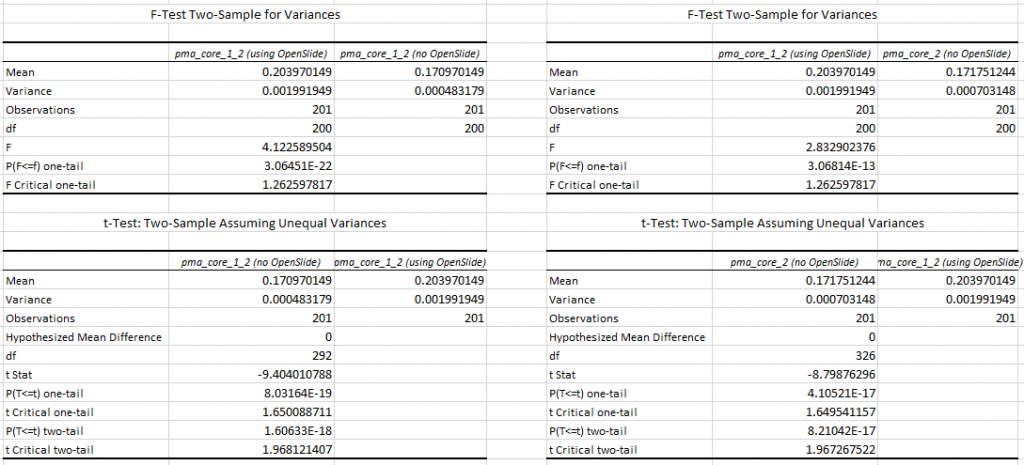
Again, we conclude that both PMA.core 1.2 and PMA.core 2 (as well as PMA.start) are faster to render Sakura SVSlide slides than OpenSlide.
What about the others?
We’re still working on getting data for the other file formats. Stay tuned!
