What is slide meta-data?
In order to know to interact with something, we first need to know what it is. Or, at least, we need you to know what it means for us. Just so we’re clear what we’re talking about.
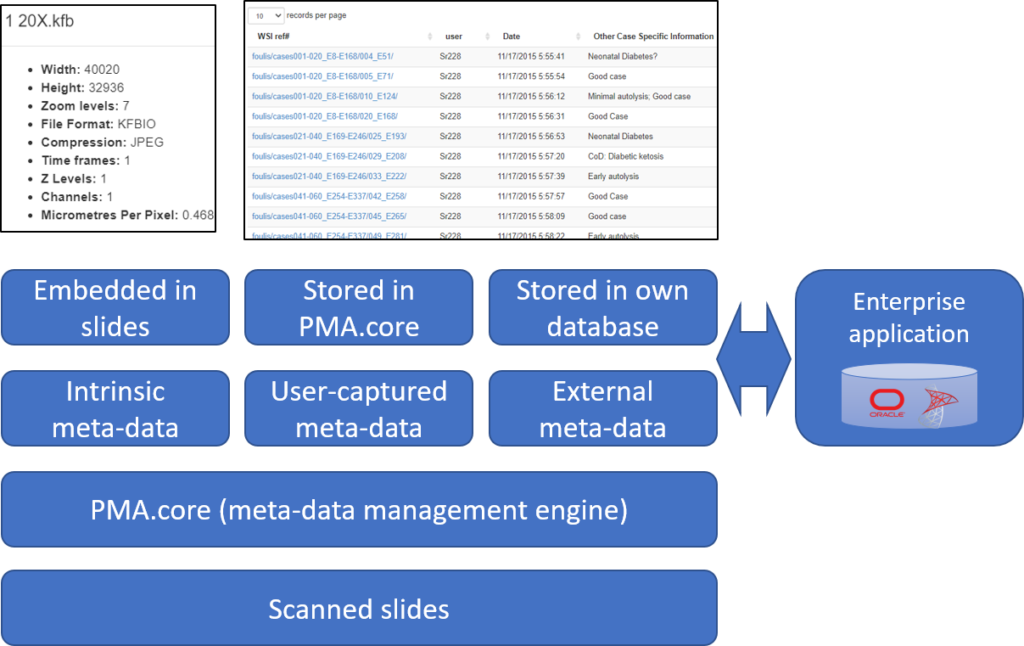
For the sake of this article, we distinguish three different kinds:
- Intrinsic meta-data: information stored within a slide’s file format. A trivial example is the slide’s pixel size, and a more advanced feature is the time it took the scanner to produce the slide.
- User-captured meta-data (forms): Pathomation’s central tile server component, PMA.core, allows for the definition of table structures. Forms are a basic data structure in PMA.core that’s picked up again and used to capture and present user-attributed data in other several Pathomation platform components, including PMA.studio, PMA.slidebox, and PMA.control.
- External data: if you have tons of external data in a separate repository and want access to it through the Pathomation software platform for digital pathology and virtual microscopy, you can link to it in real-time.
Below you can find an overview of what kind of meta-data is supported by what component and in what capacity
| Slide info | Forms | External data | |
| PMA.start | Read | Not supported | Not supported |
| PMA.core | Read | Define / read | Read |
| PMA.view | Read | Not supported | Not supported |
| PMA.studio | Read | Read / write | Read |
| PMA.slidebox | Read | Read / write | Read |
| PMA.control | Read | Read / write | Read |
Slide info
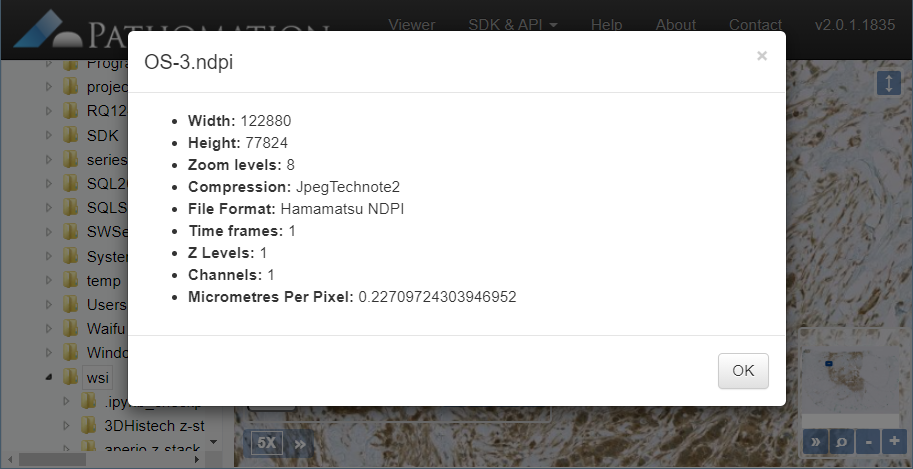
Intrinsic slide meta-data can be shown in PMA.start by clicking on the filename in the viewport.
In the upcoming versions of both PMA.view and PMA.studio, we provide a separate info-panel for this.
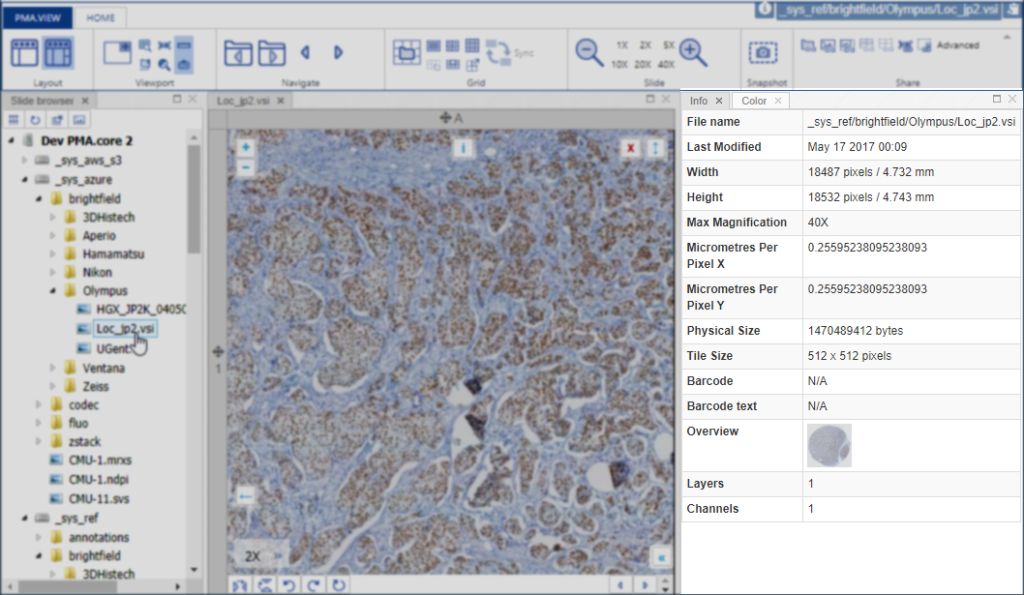
There can be more slide info available than is actually shown in our various front-end interfaces. Some information is really specific, too specialized, or irrelevant even to show to most users. Other fields are scanner-specific and don’t make sense to include on a systematic basis.
At the API and SDK level, we provide dedicated SlideInfo calls that return full hierarchical dictionary structures that do give exhaustive slide information and can be consumed as you see fit for your specific application or workflow.

Data with a twist
Forms are defined in PMA.core through the form editor.
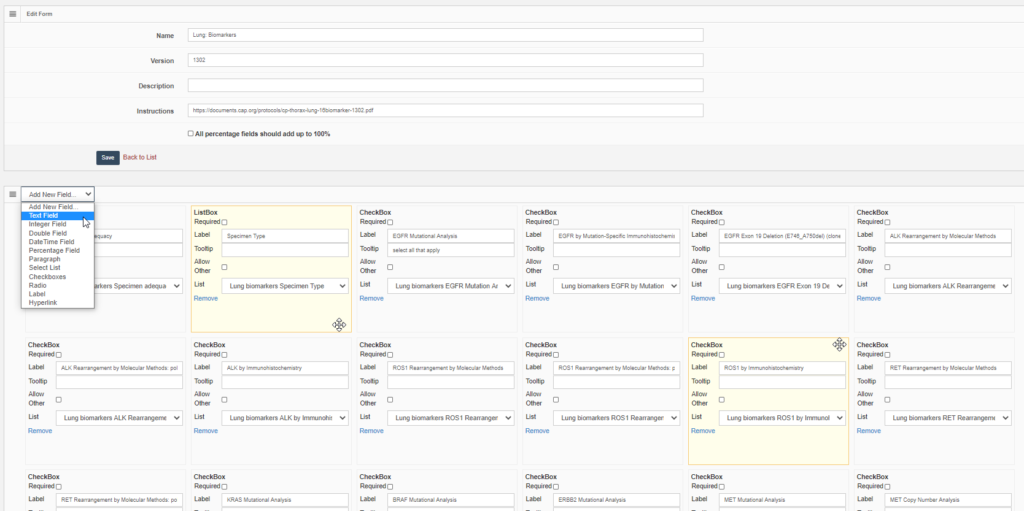
PMA.core’s forms support trivial datatypes like text or numbers (of course). We also offer scientifically relevant twists on traditional data capture. An example is that numerical data fields allow the option to be recorded as “below detectable limit”, since it’s very hard to prove that something is not present in a sample. Oftentimes, a numerical zero just means that our detection apparatus just lacks the sensitivity the unmask the presence of a specific phenomenon.
What can you do in forms? Pretty advanced things. We can model the CAP-recommended cancer protocols as PMA.core forms.
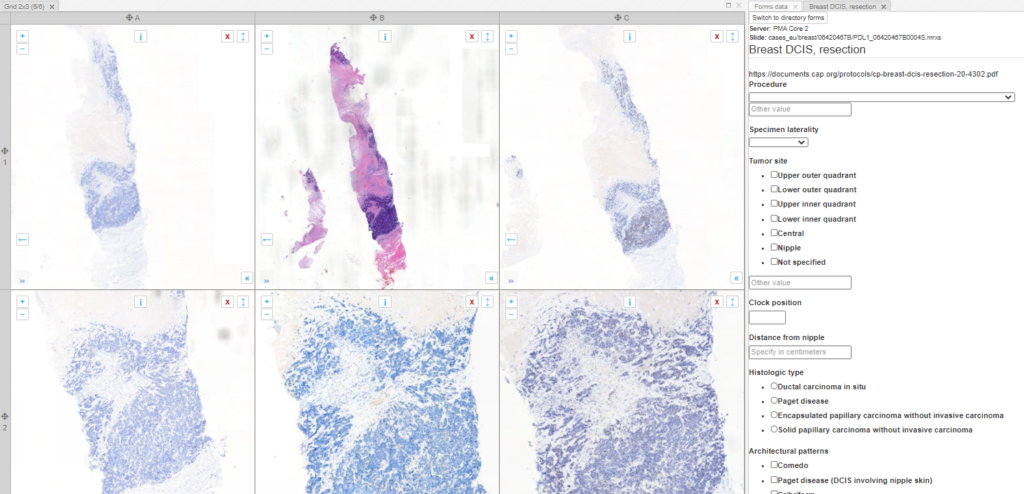
PMA.core offers different ways to interact with forms, but it doesn’t provide a data entry module. That’s because it doesn’t really make sense to enter data at this level. Data entry is done in the context of an application or workflow. Our upcoming PMA.studio will support data entry, and PMA.control offers several interaction modes. Underneath, PMA.control interaction modes rely on data stored as PMA.core forms.
You can ask PMA.core to generate a spreadsheet template based on a select folder of slides and a particular already defined form.
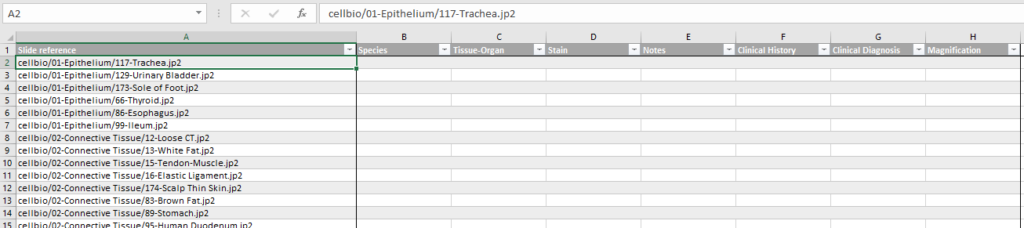
At Pathomation, we pride ourselves to be a truly open platform, so PMA.core offers several data formats to export captured form data to, including CSV, XML, and ARFF.
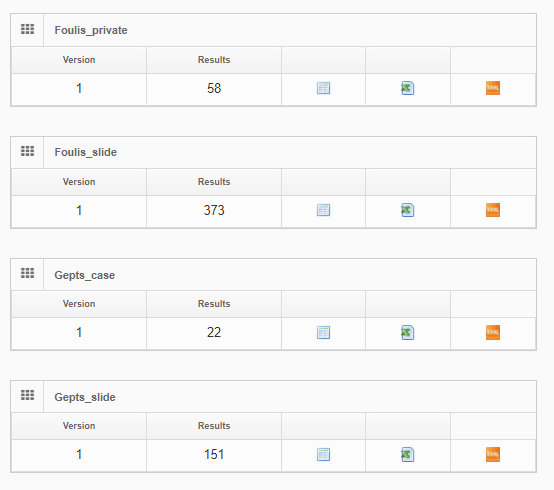
Use cases for external data
Imagine that you organize a toxicology experiment with a rodent population. In a separate database, you’ve been keeping data about each specimen’s vital statistics, dietary and behavior observations, as well as their phenotypic expression and genotypic make-up. Observations happen daily, so that’s 1750 new records per week.
You take weekly biopsies of the animals to monitor their response to a new drug you’re testing. The biopsies are prepared and stained in triplicate (good for a total of 750 new slides per week), and each slide has a barcode that can be used to trace back to the original individual animal. Your slide scanner takes care that the barcode is encoded in the slide’s filename.
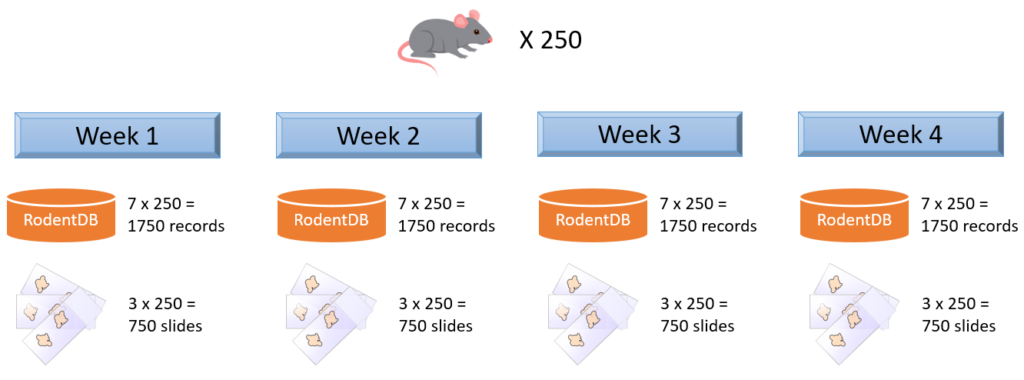
Both the slide population and the separate database keep evolving. Replication between your database and a PMA.core form is considered at some point, but deemed inefficient and error-prone, because we are talking about experimental and evolving data structures here.
The solution is to define an external data source in PMA.core. This goes in two steps: first a connection string is defined to connect to the database server.
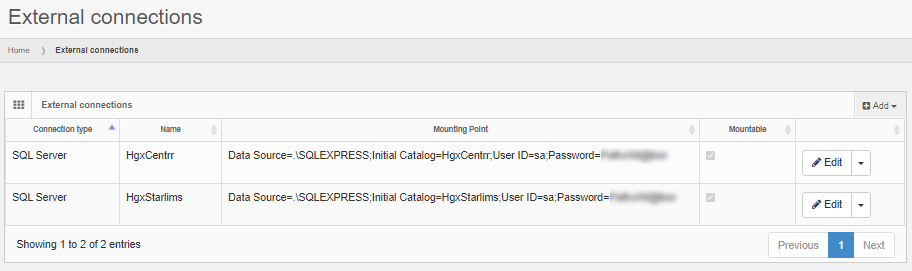
Next, the external connection is used to formulate any number of queries against.
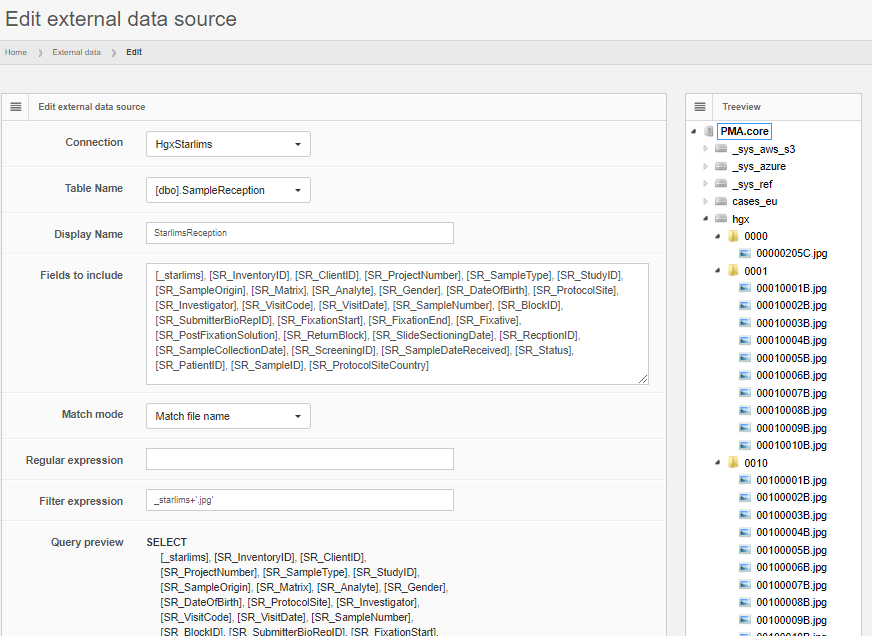
Data can be previewed within PMA.core, and subsequently is automatically propagated to other environments like PMA.studio.
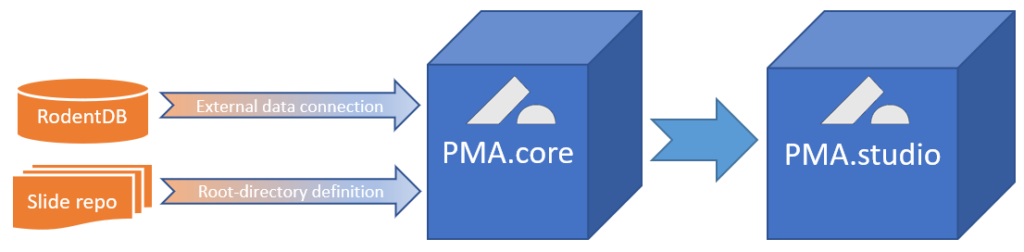
External data are everywhere. They can be in proprietary databases as in our example above, but they can also be in a (AP)LI(M)S, VNA, PACS, or EHR system. In all those cases, replication is hard and impractical at best, and can even lead to data inconsistencies and errors at worst.
The Pathomation platform for digital pathology and virtual microscopy allows for a more elegant solution.
